
|
| Polina Golland, PhD Core PI |
Our Publications
The Anatomical Variability Core develops computational models of anatomical heterogeneity in large populations for the purpose of providing accurate priors for atlas-based segmentation of anatomical structures in neuroimaging. Segmentation is particularly challenging if the shape of the anatomical structure of interest varies substantially in a population, such as would be observed in a progressive neurodegenerative disorder or cancer. Building computational models that capture highly variable anatomy and using the information to improve image segmentation are the main objectives of this Core. The goal is to develop a new generation of robust segmentation tools capable of incorporating knowledge about pathology-induced anatomical variability. Such tools are essential to achieving progress in the patient-specific analysis of disease. The work of this core is organized around the following specific aims.
- Develop Models of Anatomical Heterogeneity in a Population to Provide Better Priors for Image Segmentation in Neuroimaging.
- Develop Computational Models of Local Anatomical Variability for Integration into Segmentation.
- Validate, Deploy, and Distribute the Segmentation Tools for Image Analysis in the Presence of Pathology.
Anatomical variability that arises as a consequence of pathology, such as the neural degeneration associated with stroke, brain tumors and ALzheimer’s disease is the clinical focus of our research, but the proposed methods will be broadly applicable to other domains of naturally occurring variability in the shape of anatomical organs, well beyond neuroimaging.
The objective of our research is three-fold. Models of anatomical heterogeneity in a population are being developed in aim 1 to improve the availability of priors for image segmentation in neuroimaging. Our approach to modeling anatomical variability treats a heterogeneous population as a collection of relatively uniform sub-populations, each of which can be represented by a single training template in the segmentation algorithm. In aim 2, we are developing methods that explicitly account for registration uncertainty in building models of anatomical variability. Our approach assumes an atlas coordinate system into which all images are mapped and in which the local models of anatomical variability are constructed. The models we develop are complementary to the global variability models in Aim 1. We will explore a number of options for constructing local variability models and integrate them into segmentation. After validating the algorithms, we disseminate them to the broad medical image computing community in the form of segmentation tools to be used for image analysis.
Featured Technologies
Shape Priors for Cerebrovascular Disease Segmentation
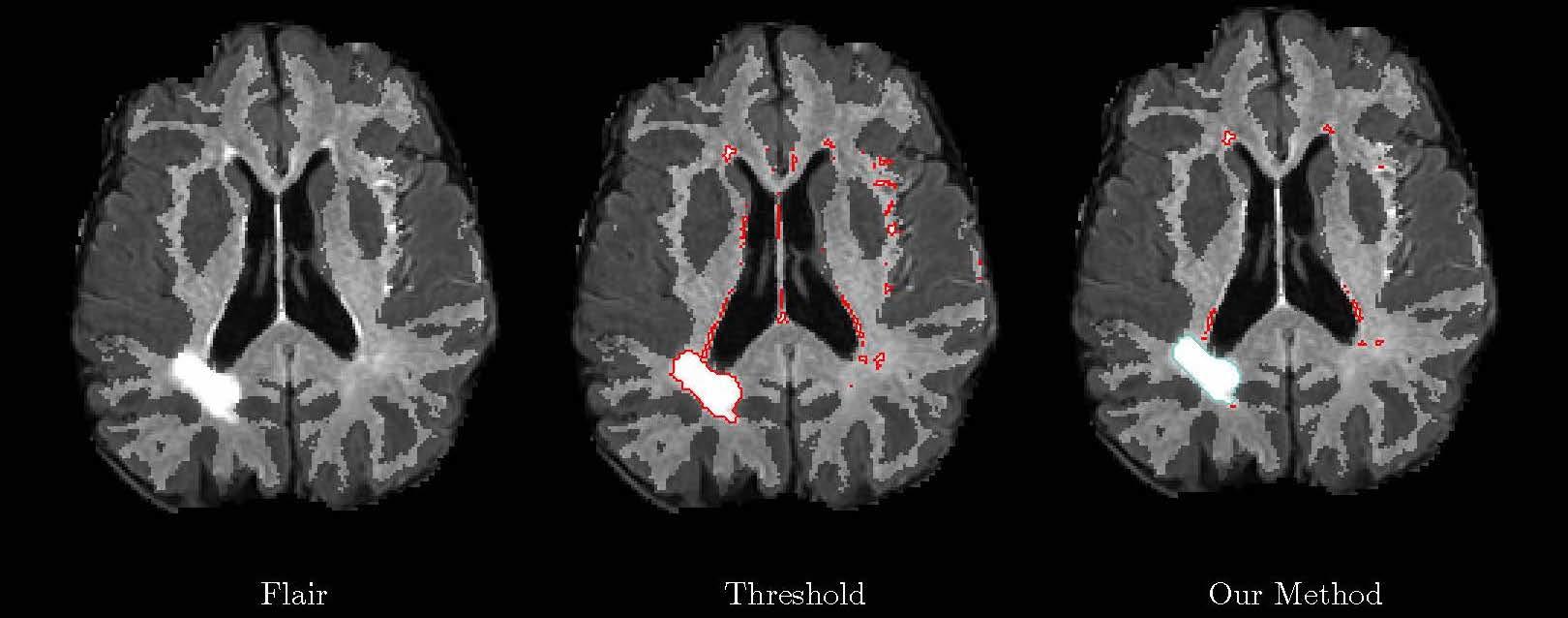
Identifying and differentiating pathologies is important for understanding the underlying mechanisms and clinical outcomes of cerebral ischemia. It is a central goal in our clinical stroke study collaboration that provides the first step in analysis and genetic association with disease. Manual delineation is infeasible in large studies of stroke that include thousands of patients. Unlike normal brain tissues and structures, the location and shape of the lesions vary significantly across patients, presenting serious challenges for prior-driven segmentation. Our generative model captures spatial patterns and intensity properties associated with different cerebrovascular pathologies in stroke patients, resulting in an inference algorithm for the automatic segmentation of cerebrovascular pathologies in clinical MR images of the brain.
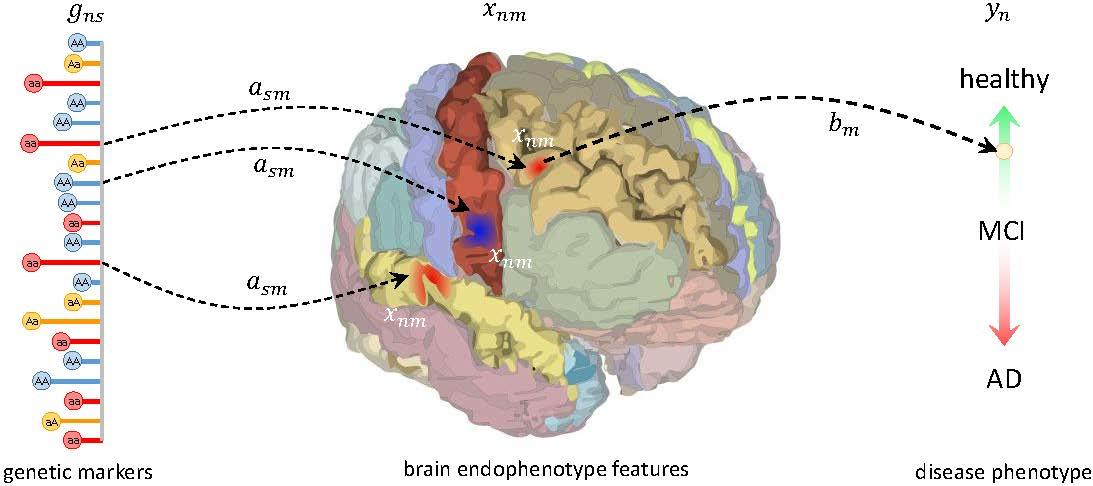
Imaging genetics studies the relationships between genetic variation and measurements from anatomical or functional imaging data, often in the context of a clinical disorder. Specifically, the use of imaging data for examining genetic associations promises new directions of analysis.
Our approach is to make use of a unified Bayesian framework for detecting genetic variants associated with disease by using image-based features as an intermediate phenotype. We jointly exploit information in the available data types, resulting in probabilistic measures of clinical relevance for both imaging and genetic markers. The resulting inference algorithm naturally handles the high dimensionality of image and genetic data to identify imaging genetic features associated with the disease.
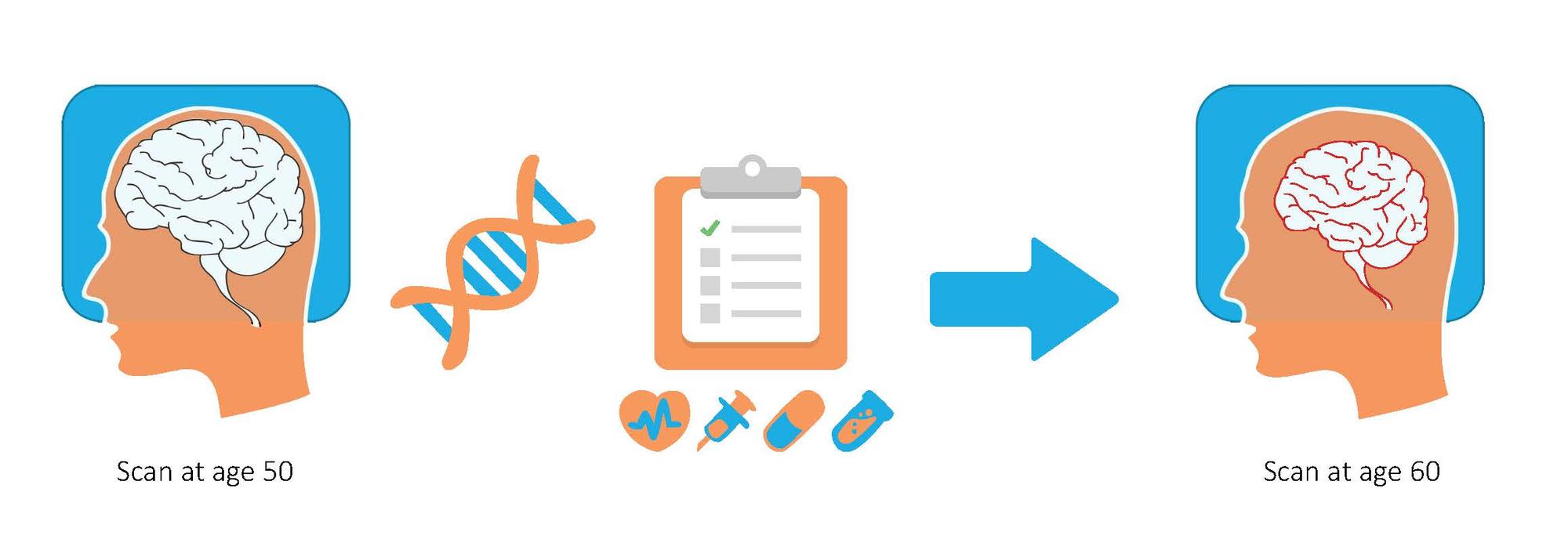
In contrast to classical genetic correlation, we aim to predict the anatomy of a patient in subsequent scans following a single baseline image, using subject-specific genetics. Such predictions promise to facilitate novel analyses in both voxel-level studies and longitudinal biomarker evaluation. Our approach involves a semi-parametric generative model that captures anatomical change through a combination of population-wide regression and the subject’s health based on individual genetic and clinical indicators. We provide prediction of follow-up anatomical scans in the ADNI cohort. We also explore novel analysis approaches that compare a patient’s scans to the predicted subject-specific healthy anatomical trajectory.
BrainPrint: Representing anatomical variability of cortical and subcortical structures
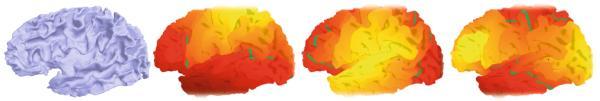
The shape information within ensembles of cortical and subcortical structures can be captured through novel representations. BrainPrint compactly represents shapes by solving the 2D and 3D Laplace-Beltrami operator on boundary (triangular) and volumetric (tetrahedral) meshes. BrainPrint is a discriminative representation that captures unique information about the subject’s anatomy. In particular, a robust classifier trained on BrainPrint representations from a database of brain scans can reliably identify the subject in a new scan. In an example dataset containing over 3000 MRI scans from the ADNI dataset, BrainPrint yields correct subject classification of a scan with 99.8% accuracy. We have also investigated applications to longitudinal studies and to a wide range of anatomical analyses commonly performed in clinical research. Computer-aided diagnosis of Alzeimer’s disease and it’s prodromal stage of mild cognitive impairment also benefits from using BrainPrint to characterize the shapes of brain structures. We derive features for automated diagnosis from BrainPrint by computing lateral shape differences and projections onto the principal components. Our approach is to perform classification using a generalized linear model that includes regularization and automatic model selection. The resulting algorithm won the second prize at the ‘MICCAI 2014 Challenge on Computer-Aided Diagnosis of Dementia Based on Structural MRI Data’.
Image Imputation
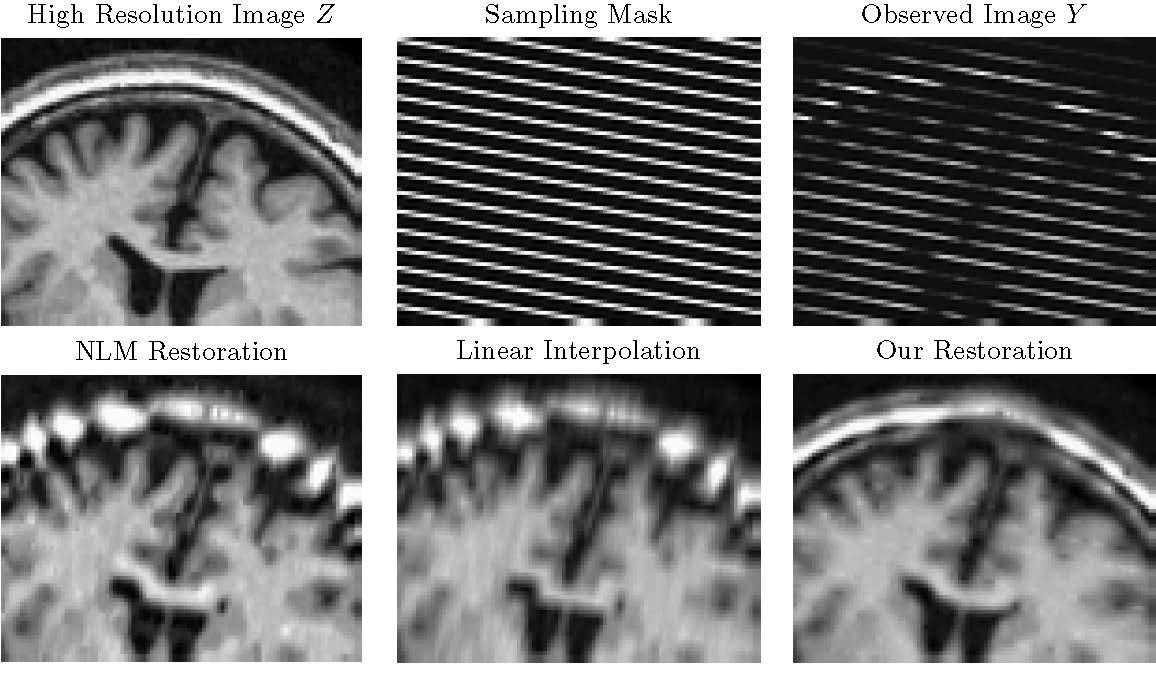
Large databases of clinical images contain a wealth of information, yet medical acquisition constraints result in sparse scans that miss much of the anatomy. These characteristics often render computational analysis impractical as standard processing algorithms tend to fail when applied to such images. Our goal is to enable application of existing algorithms that were originally developed for high resolution research scans to significantly undersampled scans. Our algorithm creates high resolution anatomically plausible images consistent with acquired clinical brain MRI scans with large inter-slice spacing. The model captures fine-scale anatomical similarity across subjects in clinical image collections. We use it to fill in the missing data in scans with large slice spacing. This method promises to facilitate subsequent analysis not previously possible with scans of this quality.
Collaborations
We maintain an active collaboration with our colleagues in the Neurology Department of Mass General Hospital with the goal of analyzing clinical brain MRI scans of stroke patients to identify genetic associations of imaging biomarkers of cerebrovascular health. We also work with the National Center for Image Guided Therapy at the Brigham and Women’s Hospital to develop robust methods for segmentation and registration of brain tumor scans for surgical navigation. We have several joint projects with the Center for Functional Neuroimaging technologies at the Martinos center for Biomedical Imaging, Massachusetts General Hospital.
Outreach and Dissemination
We make our algorithms available to the broad medical image computing community by integrating them into open source software platforms such as 3D Slicer and releasing the code on github. When the computational solutions mature, we also work with our collaborators to integrate them into the clinical environment.
Our contributions to the medical image computing community go beyond scientific publications and associated open source implementations.. The members of the Core have organized and chaired the Brain Tumor Segmentation Challenge (BraTS) at MICCAI, the MICCAI Imaging Genetics Workshop, and the main MICCAI Conference. As part of NAC, we host the annual winter project weeks at MIT, where computer scientists, software developers, clinicians, and clinical researchers to work together towards better open source solutions to medical image computing.
Interactions with Other Cores
We collaborate closely with the Spatio-Temporal Modeling Core on feature-based anatomical models and on high dimensional inference from image data, with joint projects, thesis co-supervision, and joint publications.
We work with the Microstructure Imaging Core and the Slicer Core in our work to support the clinical collaborators in the National Center for Image Guided Therapy.
