OBJECTIVES: Disrupted auditory networks play an important role in the pathophysiology of psychosis, with abnormalities already observed in individuals at clinical high-risk for psychosis (CHR). Here, we examine structural and functional connectivity of an auditory network in CHR utilising state-of-the-art electroencephalography and diffusion imaging techniques.
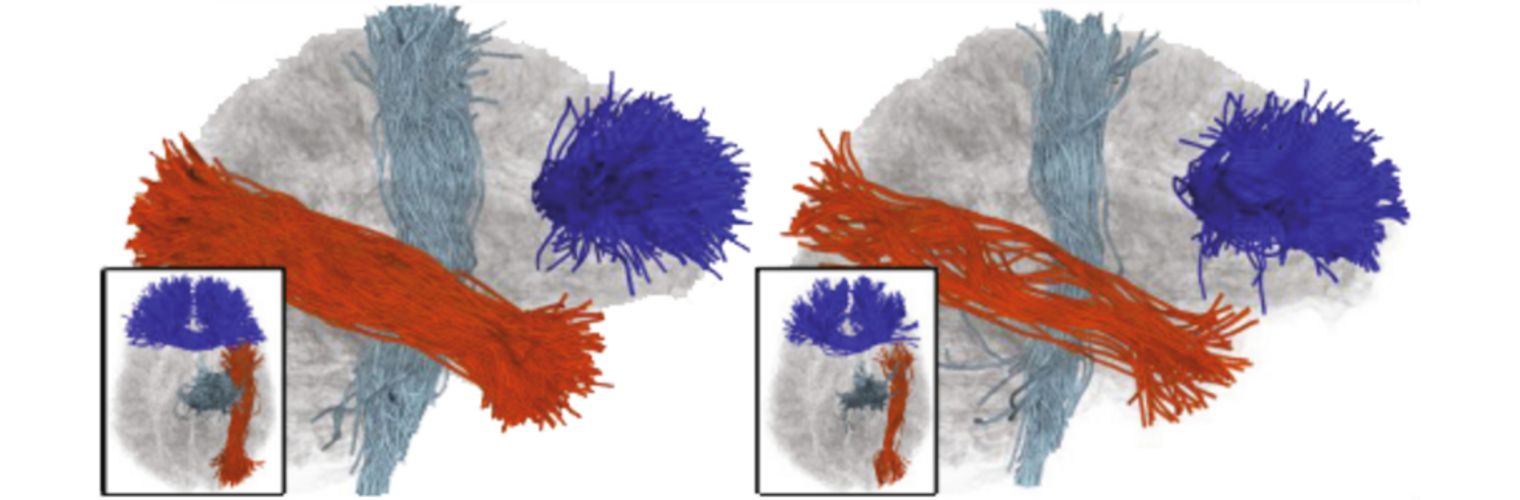
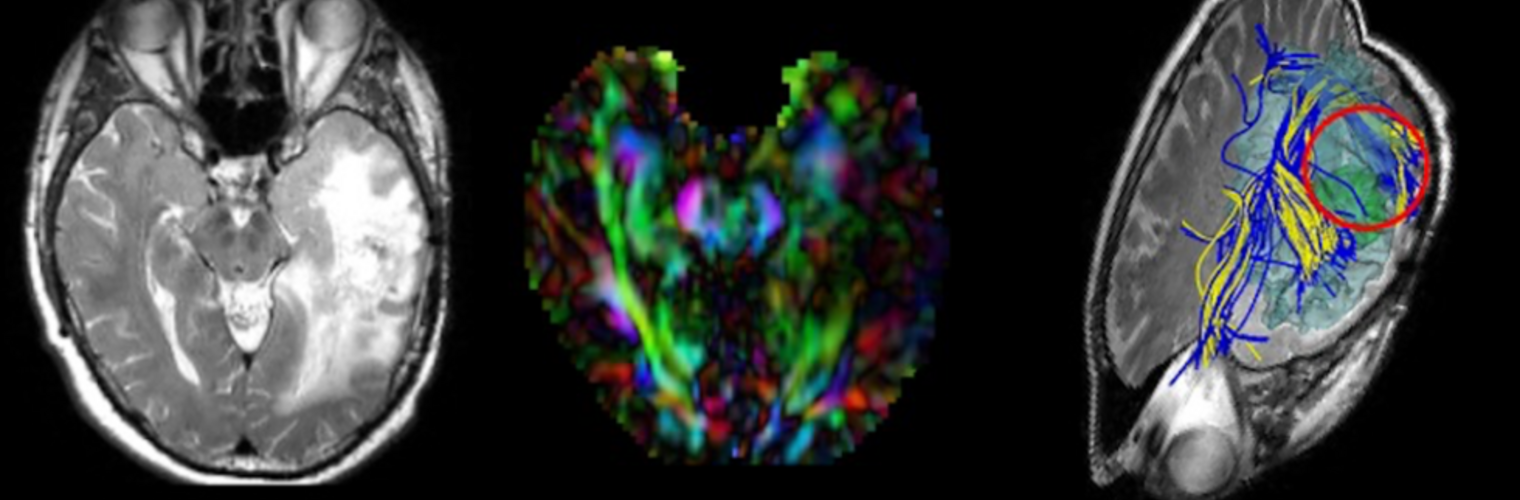
METHODS: Twenty-six CHR subjects and 13 healthy controls (HC) underwent diffusion MRI and electroencephalography while performing an auditory task. We investigated structural connectivity, measured as fractional anisotropy in the Arcuate Fasciculus (AF), Cingulum Bundle, and Superior Longitudinal Fasciculus-II. Gamma-band lagged-phase synchronisation, a functional connectivity measure, was calculated between cortical regions connected by these tracts.
RESULTS: CHR subjects showed significantly higher structural connectivity in the right AF than HC (p < .001). Although non-significant, functional connectivity between cortical areas connected by the AF was lower in CHR than HC (p = .078). Structural and functional connectivity were correlated in HC (p = .056) but not in CHR (p = .29).
CONCLUSIONS: We observe significant differences in structural connectivity of the AF, without a concomitant significant change in functional connectivity in CHR subjects. This may suggest that the CHR state is characterised by a decoupling of structural and functional connectivity, possibly due to abnormal white matter maturation.