
|
| Sandy Wells, PhD Core PI |
Our Publications
The Spatio-Temporal Modeling Core develops statistical methods for the analysis of functional MRI (fMRI) in populations and for the group-wise registration of large datasets. New lines of research include the analysis of multi-modal functional data and studies of the relationship between structure and function, which are used for neurosurgical intervention and other applications that require detailed maps of cortical areas that show these relationships. The work of this core is organized around the following specific aims.
1. State Space Modeling (SSM) Approach for fMRI Analysis
2. Feature-Based Analysis (FBA) of Structural MRI
3. Methodology for Quantitative Magnetic Susceptibility Mapping (QSM)
The methods developed in the specific aims will be applied in a variety of clinical scenarios.
Under Aim 1, we are adapting current SSM methods to incorporate hierarchical state sequences. The latter technology will be applied in a clinical study of cognitive dysfunction in patients with HIV infection and/or histories of methamphetamine use with our clinical partner, TMARC, centered at the University of California, San Diego. Under Aim 2, we are continuing to refine our current algorithms for feature-based analysis of structural MRI in groups of subjects and the resulting FBA technology will be applied in a pilot project that analyzes historical multiple sclerosis (MS) data. Under Aim 3, we are extending current methods for QSM to analyze tissues from MRI phase data in healthy subjects. This data will then be applied in at least two clinical projects, Parkinson's Disease (3A) and Traumatic Brain Injury (3B). The scientific team is composed of experts in mathematics, statistical modeling, image analysis, and functional (fMRI) and structural magnetic resonance imaging (MRI).
Using state-space model (SSM) as a platform for aim 1, we are developing a framework that consists of statistical measures, software, experimental designs, and neuroscientific interpretations. This new framework will be used to study the spatial character of mental activity, its temporal structure, and its implications on the organization of the mental processes of an individual and their salient differences between populations. By using dynamic multivariate methods that study the spatio-temporal entirety of mental activity to further our understanding of the neurobiological underpinnings of complex psychological conditions, we will elucidate the temporal ordering of neural cascades, and in this way improve the possibility of detecting abnormal spatio-temporal pathways in mental processing due to biological and psychological pathologies, with implications on cognitive and preclinical studies in neurology and psychiatry pertaining to, e.g., disease, aging, and dementia. Moreover, identifying the salient layers of human cognition and the interactions between them will enable the neurobiological investigation of the complex cognitive, cultural, and social factors in neuroscientific experiments.
Developments in feature extraction (aim 2) will focus on identifying classes of image features most effective for feature-based analysis (FBA) of MS. Experiments will involve dual echo MR image data, consisting of proton density and T2-weighted brain images for each subject. How best to extract and combine features from multiple image modalities into an FBA framework remains an open research question. Research will compare the result of FBA from features extracted independently in different image modalities vs. extraction in joint representations. Furthermore, different feature classes developed for 2D images will be extended to volumetric data and assessed in terms of effectiveness in FBA. Different modalities such as PD and T2 offer complementary information regarding the underlying subject anatomy, and extracting features from a joint image representation combining modalities may prove more effective than independently in individual modalities.
Patients with Parkinson's disease (PD) suffer a 60% to 70% reduction of neurons in the substantia nigra (SN) by the time the first clinical symptoms of PD are noticed by the patient, precluding effective neuroprotective therapies. It is possible that neuroprotective medications would be effective if they could be initiated before there was substantial cell loss, which would require a noninvasive method to detect preclinical markers of disease. Hypoechogenicity on transcranial sonography has been identified as a potential biomarker of SN injury. The level of hypoechogenicity in PD patients, however, is highly variable (range 68% to 99%). A study of 20 post mortem PD brains found a positive correlation between iron concentration and echogenicity in the SN, suggesting imaging methods sensitive to iron concentration should allow robust identification of the SN. We are developing a new approach to QSM in aim 3, termed atlas-based susceptibility mapping (ASM), with the goal of classifying Parkinson's disease patients and controls based on mean susceptibility estimates in the SN.
Featured Technologies
1. State Space Modeling (SSM) Approach for fMRI Analysis
In contrast to localization-based methods, we use a multivariate dynamical model to represent the brain transitioning through an abstract state-space as it performs a mental task. Here, for example, we demonstrate the presence of sequences of well-defined states in fMRI recordings, each corresponding to characteristic distributions of metabolic activity in the brain and highly correlated with the perceptual, cognitive, or affective mental state (i.e., “brain state”) of the subject [Janoos, 2013].
2. Feature-Based Analysis (FBA) of Structural MRI
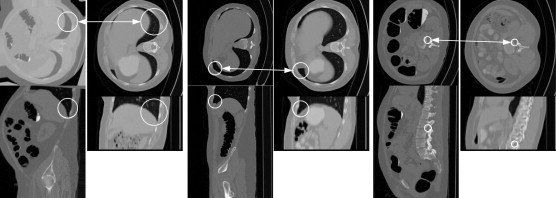
Feature-based analysis (FBA) is a recent technique in which data are represented as a set of distinctive local features or patches rather than the more traditional image voxels. Here, we show feature-based alignment (FBA), a general method for efficient and robust model-to-image alignment. Volumetric images, e.g., CT scans of the human body, are modeled probabilistically as a collage of 3D scale-invariant image features within a normalized reference space. The example shows model-subject correspondences in CT volumes. Each image consists of upper (axial) and lower (sagittal) image slices. White circles illustrating the location and scale of features in the ACRIN model (left) and in the new subject (right). The arrows indicate corresponding features [Toews, 2012].
3. Methodology for Quantitative magnetic Susceptibility Mapping (QSM)
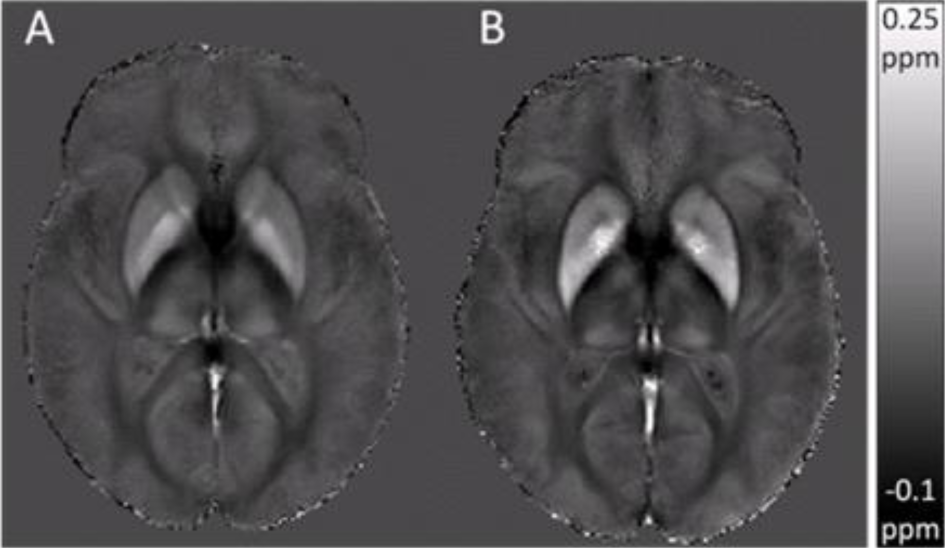
The image demonstrates the influence of age on iron accumulation in the brain, using a new approach to Quantitative magnetic Susceptibility Mapping (QSM), termed atlas-based susceptibility mapping (ASM). The ASM group averages for young (A) versus elderly (B) subjects show an age-dependent increase in estimated susceptibility values in subcortical regions known to accumulate iron in normal aging. Excessive iron accumulation occurs in a range of neurodegenerative disorders, for example, Parkinson’s disease (PD), Alzheimer’s disease (AD), Multiple sclerosis (MS), and Huntington’s disease (HD), as well as in traumatic brain injury. These iron deposits cause spatial variations in the magnetic susceptibility of tissue, which result in magnetic field perturbations that alter the phase of the MRI signal. Reconstructing magnetic susceptibility maps from MRI phase data can provide valuable insights into iron distribution in vivo.
In contrast to localization-based methods, we use a multivariate dynamical model to represent the brain transitioning through an abstract state-space as it performs a mental task. Here, for example, we demonstrate the presence of sequences of well-defined states in fMRI recordings, each corresponding to characteristic distributions of metabolic activity in the brain and highly correlated with the perceptual, cognitive, or affective mental state (i.e., “brain state”) of the subject [Janoos, 2013].
Feature-based analysis (FBA) is a recent technique in which data are represented as a set of distinctive local features or patches rather than the more traditional image voxels. Here, we show feature-based alignment (FBA), a general method for efficient and robust model-to-image alignment. Volumetric images, e.g., CT scans of the human body, are modeled probabilistically as a collage of 3D scale-invariant image features within a normalized reference space. The example shows model-subject correspondences in CT volumes. Each image consists of upper (axial) and lower (sagittal) image slices. White circles illustrating the location and scale of features in the ACRIN model (left) and in the new subject (right). The arrows indicate corresponding features [Toews, 2012].
Research Highlights
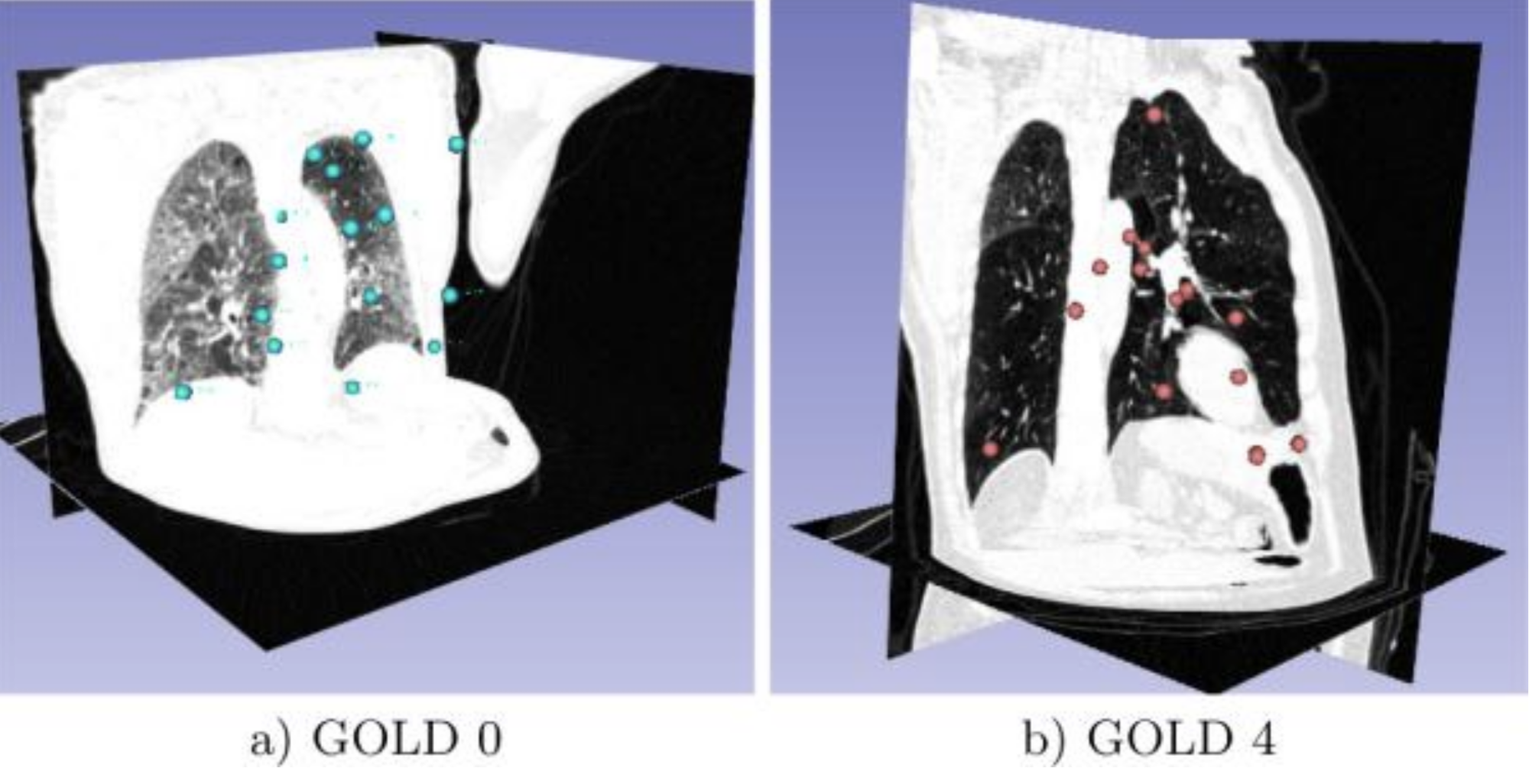
Invariant Feature-Based Analysis (FBA) of Medical Images During this year Toews and Wells presented their overview of an inference method that is well-suited to large sets of medical images based upon a framework where distinctive 3D scale-invariant features are indexed efficiently to identify approximate nearest-neighbor (NN) feature matches in O(log N) computational complexity in the number of images N. Shown here are Lung CT visualizations of the 20 disease-informative features of COPD corresponding to Gold 0 (a) and Gold 4 (b) disease severity [Toews, 2016].

Keypoint Transfer Segmentation An image segmentation method is presented that transfers label maps of entire organs from the training images to the novel image to be segmented. Coronal views of example segmentation results for contrast-enhanced CT (left) and whole body CT (right) are overlaid on intensity images. Each series reports segmentations in the following order: manual, key point transfer, and locally weighted multi-atlas [Wachinger, 2015].
Developments in State Space Modeling (SSM) of fMRI under Aim 1
The Wells science team reported the following progress in reference to aim 1. State Space Modeling Wells and Toews visited colleagues at the University Hospital Geneva and gave a talk summarizing their state space analysis methodology [Wells 2015]. The Geneva group is prominent in the state-space analysis of EEG, and they are currently collecting simultaneous EEG and fMRI of epilepsy patients. They discussed potential approaches to state-space analysis of such joint data. The other progress under this aim applies to the application of SSM of fMRI in a collaboration with Prof. Greg Brown and the NIH-Funded Translational Methamphetamine Research Center (TMARC) at UCSD. For progress related to this collaboration (see TMARC CP, below).
Developments in Feature Based Analysis (FBA) of medical Images under Aim 2
The Feature Based Analysis (FBA) project has made significant progress and is producing outstanding results. FBA generates hundreds to thousands of features per scan for MRI and CT data. To date, the application of the technology includes segmentation, registration, neuroimage analysis, and disease analysis. In this year, two publications appeared related to progress noted in the last report, one on COPD disease classification [Toews, 2015] and a second on a segmentation method called Keypoint Transfer Segmentation [Wachinger, 2015]. In yet another collaborative effort, the FBA methodology was used to register 3D cine ultrasound images of the heart, leading to a manuscript submission [Bersvendsen, 2016]. The team also used feature correspondence counts to define a Jaccard distance between subjects and showed reliable detection of twins in data from the human connectome project. By now, the FBA technology developed in NAC has been successfully applied in diverse applications; the emergent theme is that FBA combined with simple algorithms enables good performance in many areas. A summary of the activity was reported in several talks.
Development in Quantitative Susceptibility Mapping (QSM) under Aim 3
Quantitative Susceptibility Mapping QSM developments let to the appearance of a journal article describing the methodology and its application to analyzing age-related changes in the magnetic susceptibility of sub-cortical nuclei [Poynton, 2015]. While most of our research has been on the analysis of fMRI task data, we have recently collaborated on state space analysis of resting state EEG data (Custo 2017).
At UCSF the technology has been applied to 7T imaging of Huntington’s Disease. (Poynton, C. B., et al. "Quantitative Susceptibility Mapping of Huntington’s Disease at 7 Tesla." Proceedings of the Joint Annual Meeting of the International Society for Magnetic Resonance in Medicine and the European Society for Magnetic Resonance in Medicine. 2014.)
Other Collaborative Projects
Tumor Associated Seizures in Glioblastoma In our service collaboration with the National Center for Image-Guided Therapy (NCIGT), we guided the development of a methodology, based on tolerance limits, for validating image registration methods in specific applications [Fedorov, 2014]. We also provided analysis advice in a local project on the relationships among tumor region, seizures, and genetics (FIGURE 2). Tumor-associated seizures (TAS) are a common and significant cause of morbidity. Both imaging and gene expression features play significant roles in determining TAS, with strong interactions between them. We describe gene expression imaging tools which allow mapping of brain regions where gene expression has significant influence on TAS, and apply these methods to study 77 patients who underwent surgical evaluation for supratentorial glioblastomas. Tumor size and location were measured from MRI scans. A 9-set gene expression profile predicting long-term survivors was obtained from RNA derived from formalin-fixed paraffin embedded tissue. A total of 32 patients (42%) experienced preoperative TAS. Tumor volume was smaller (31.1 vs. 58.8 cubic cm, p<0.001) and there was a trend toward median survival being higher (48.4 vs. 32.7 months, p=0.055) in patients with TAS. Although the expression of only OLIG2 was significantly lower in patients with TAS in a groupwise analysis, gene expression imaging analysis revealed regions with significantly lower expression of OLIG2 and RTN1 in patients with TAS. Gene expression imaging is a powerful technique that demonstrates that the influence of gene expression on TAS is highly region specific. Regional variability should be evaluated with any genomic or molecular markers of solid brain lesions.
Gaussian Random Field Theory In collaboration with colleagues at MIT, in addition to (Wachinger 2015), we applied Gaussian Random Field theory to interpolating MRI images [Wachinger, 2014 MICCAI). We also contributed, with colleagues in the UK, on the development of probabilistic methods for segmenting the neocortex of a population of subjects, driven by diffusion MRI [Parisot, 2015].
Magneto Hydro Dynamics The TRDs collaboration with Children’s Hospital Boston on epilepsy image analysis saw the submission of an article on animal validation of direct MRI observations of tissue magnetic fields due to neural activity. We investigated an alternative mechanism, magneto hydro dynamics, that could explain the observations in humans [Balasubramanian, 2015].
