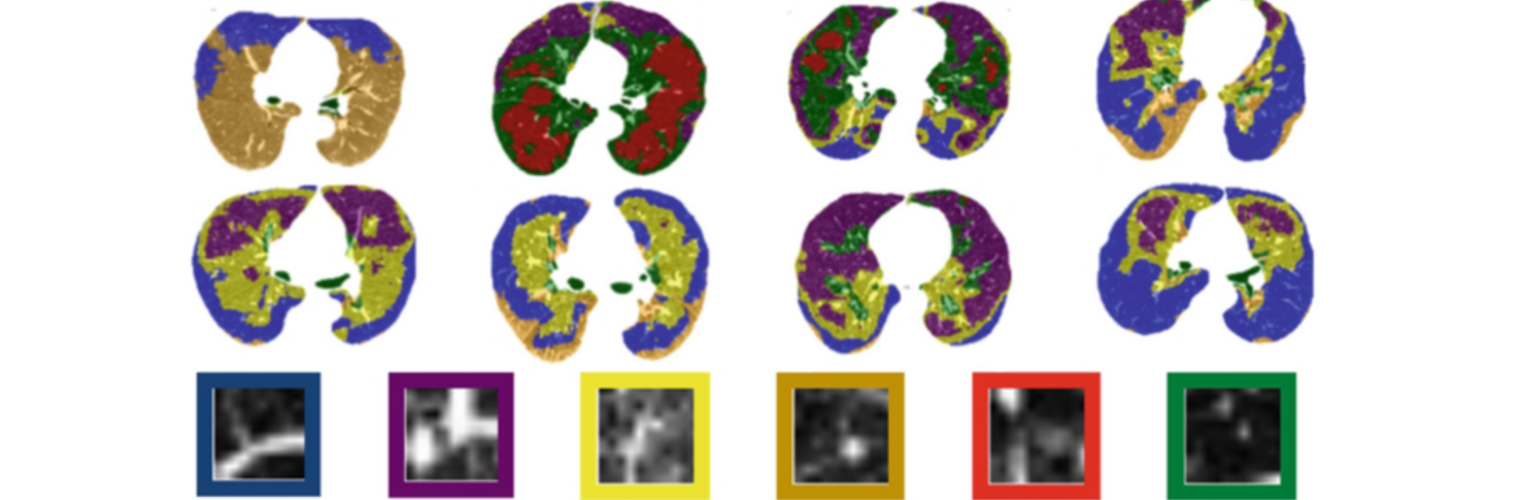
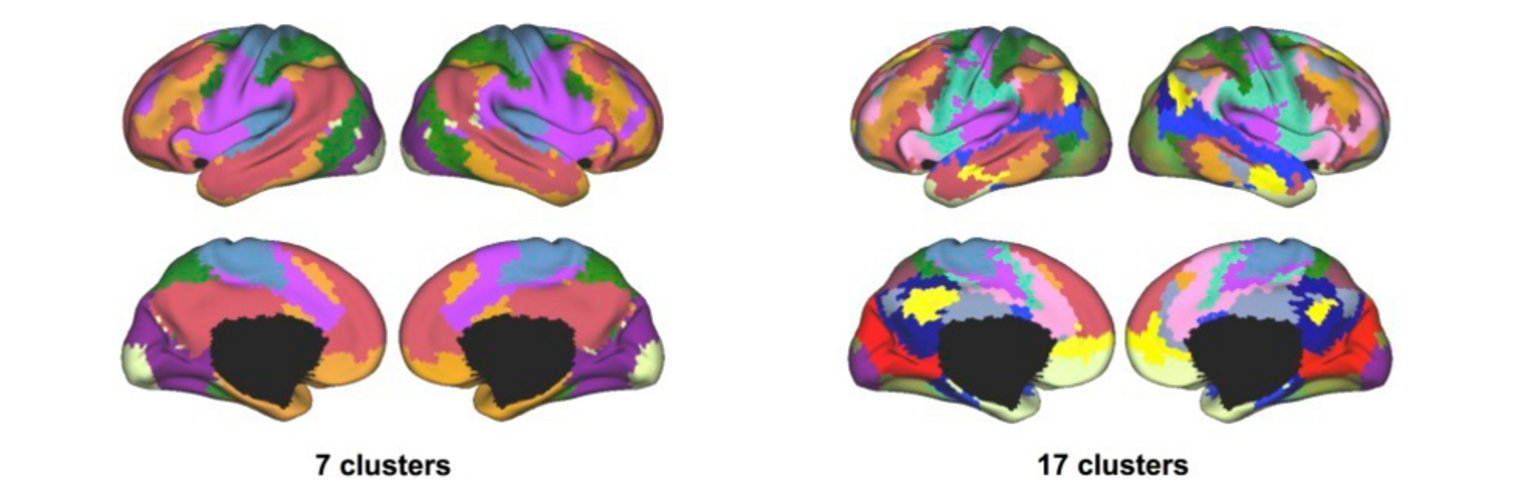
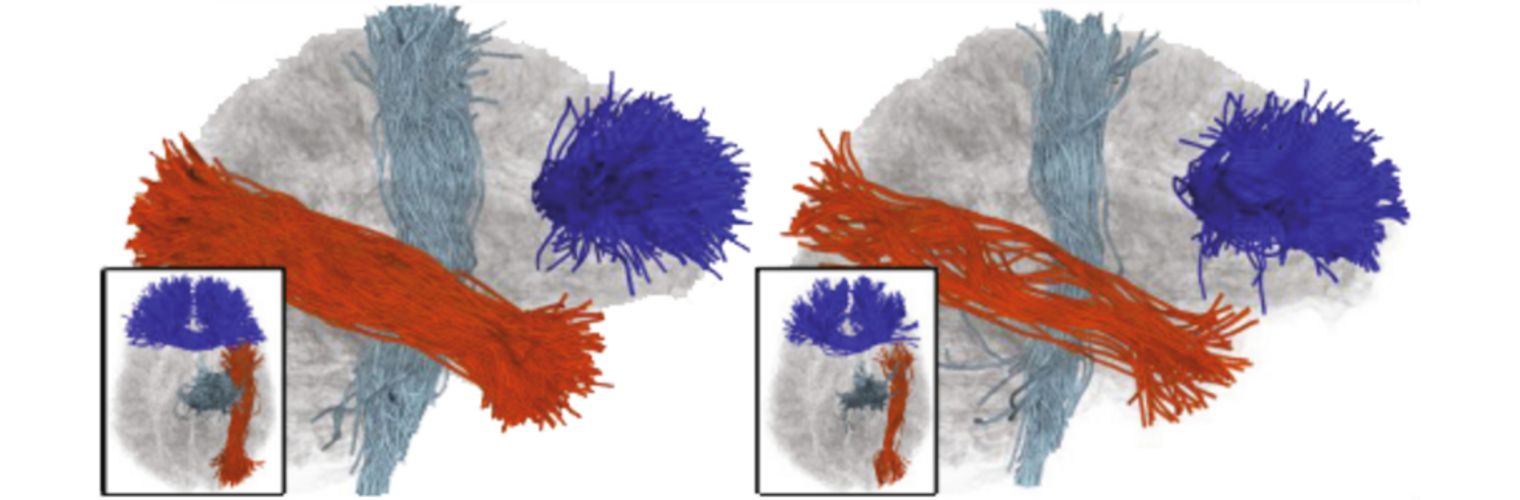
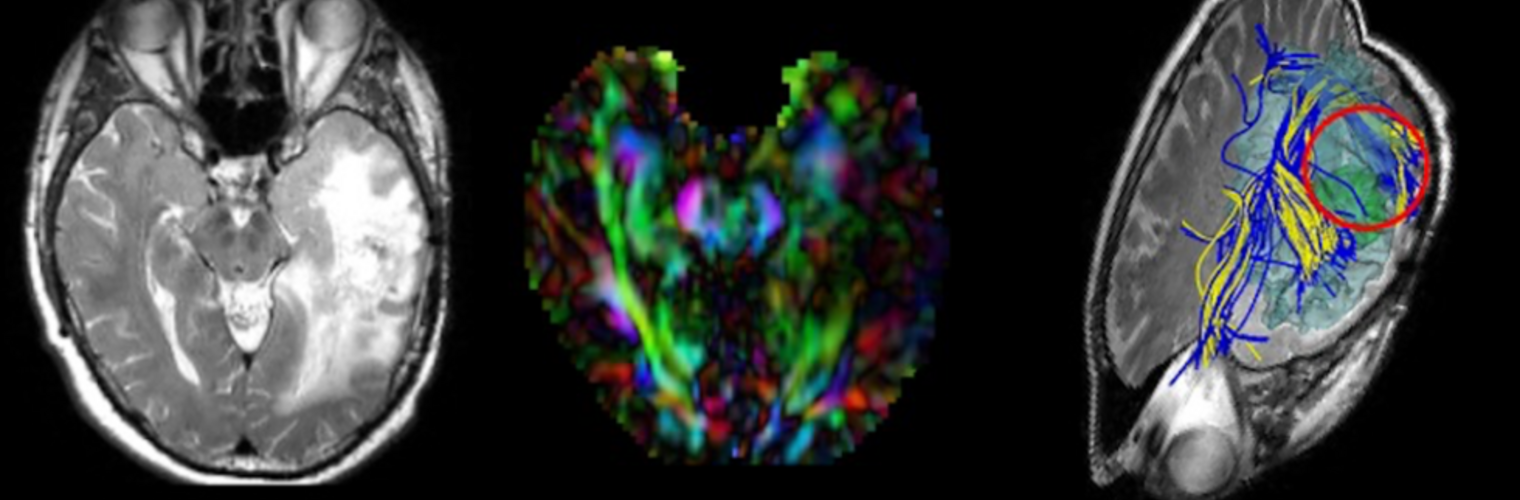
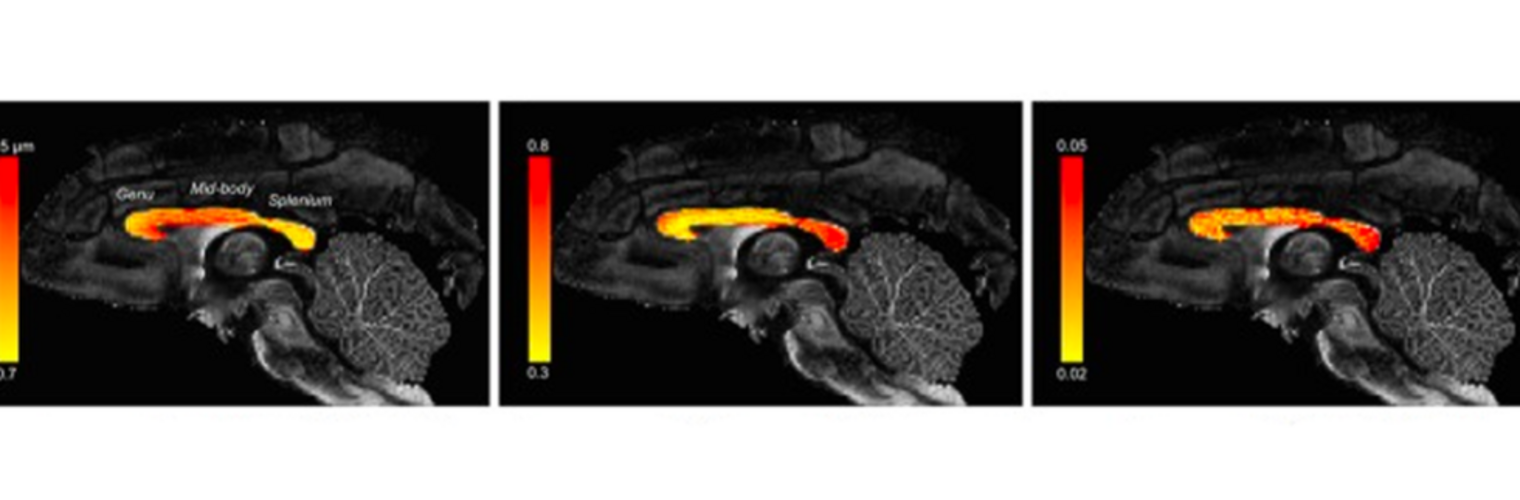
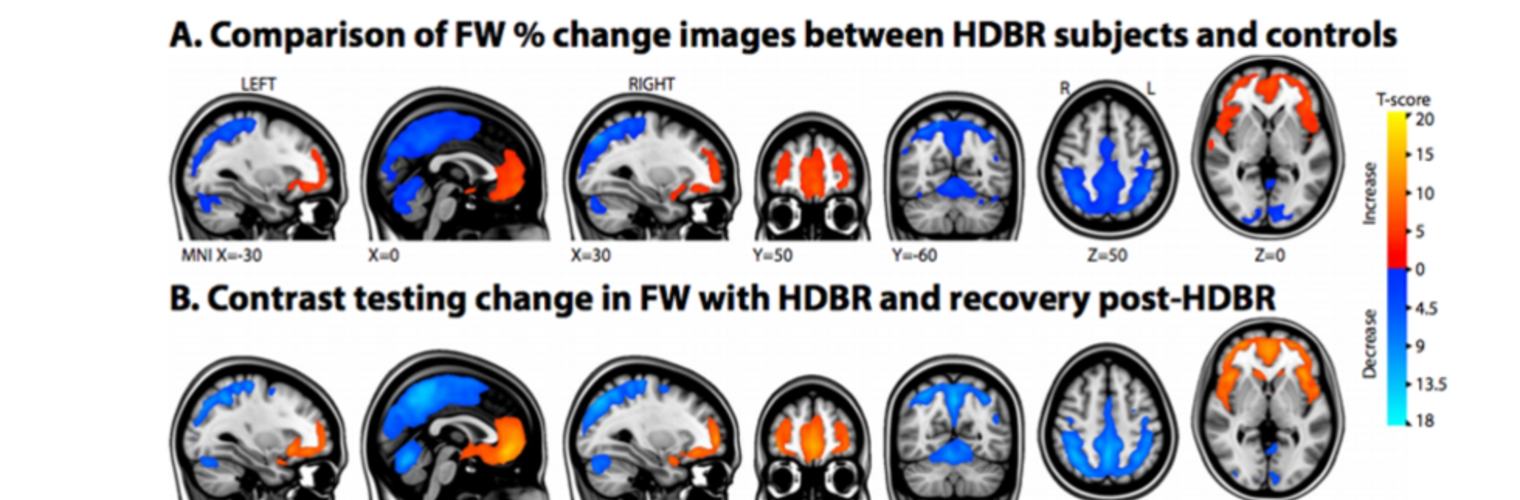
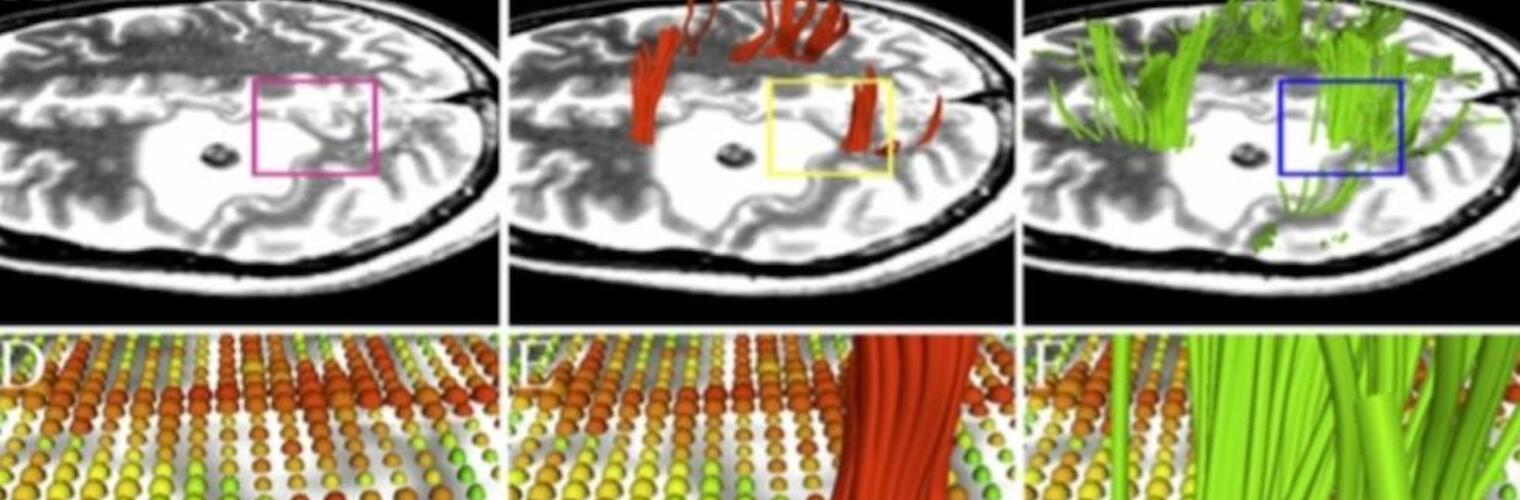
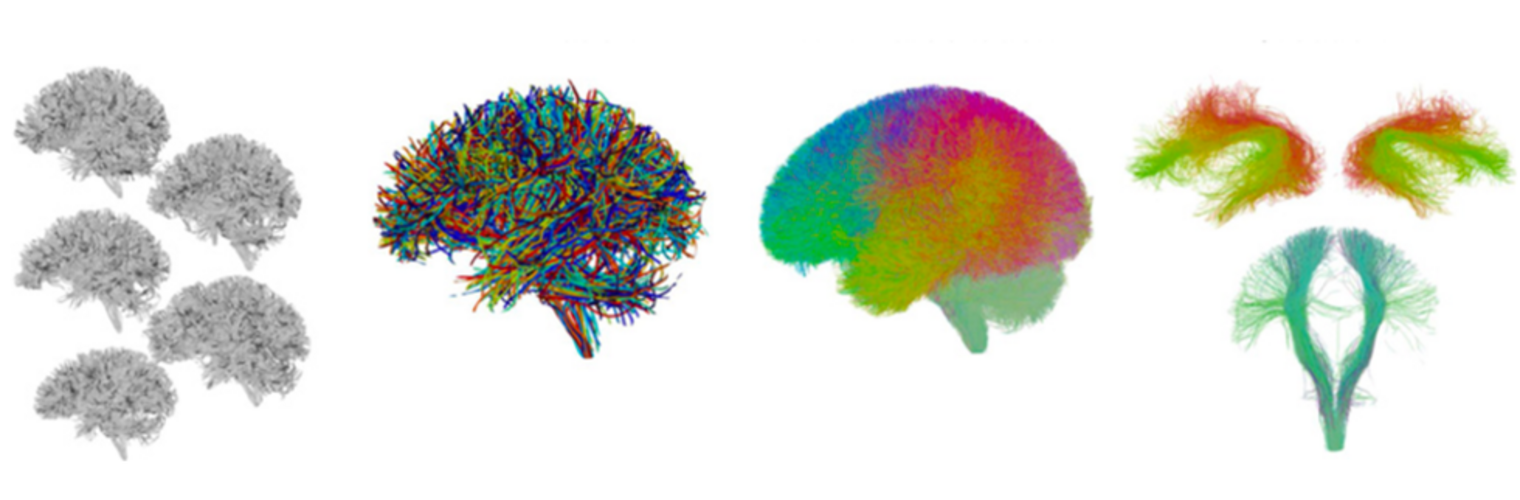
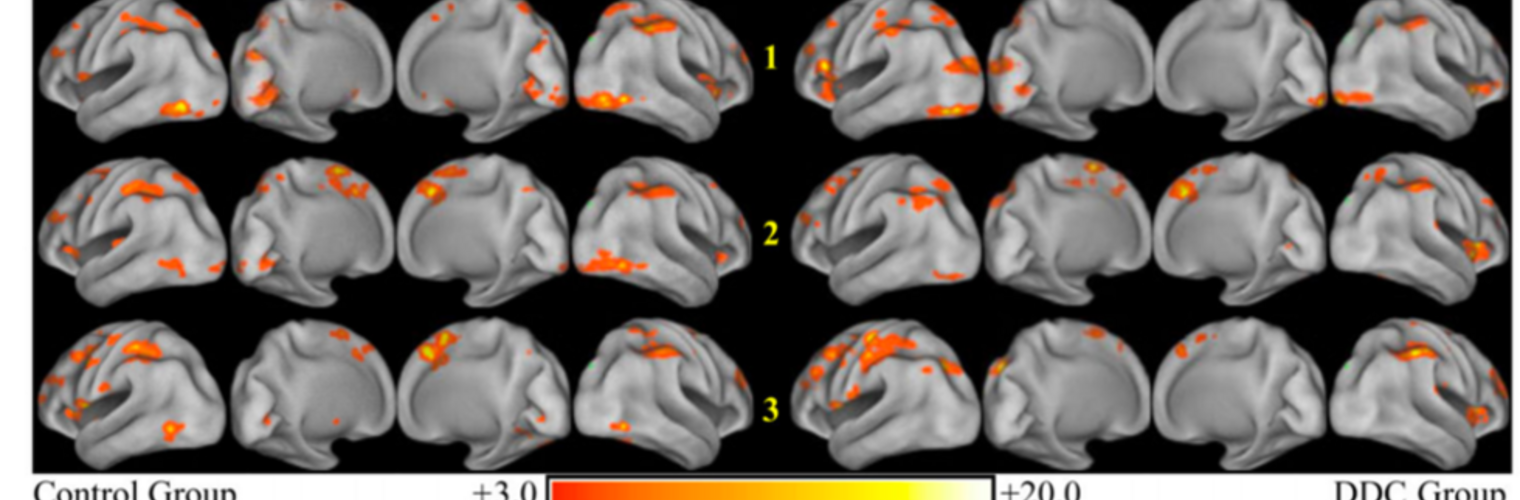
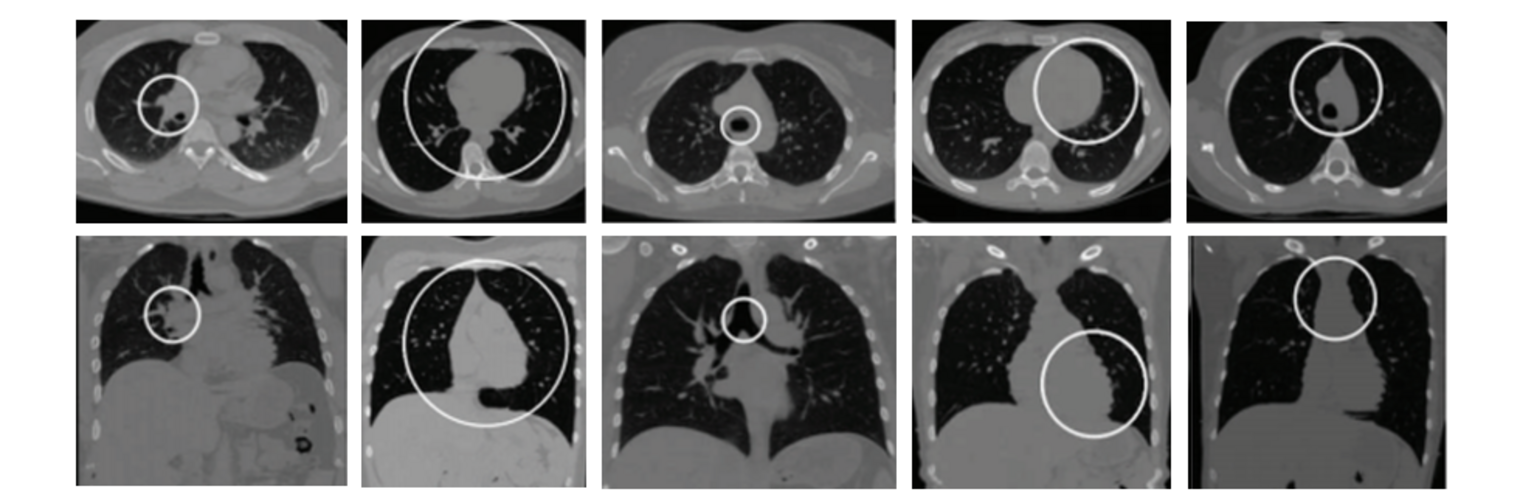
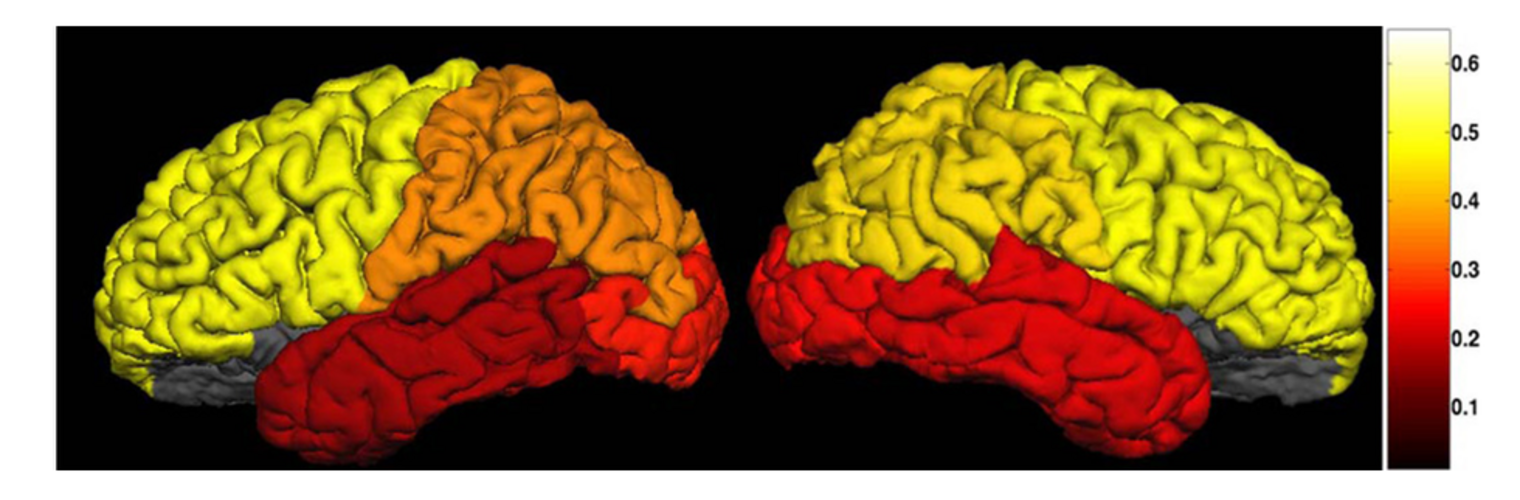
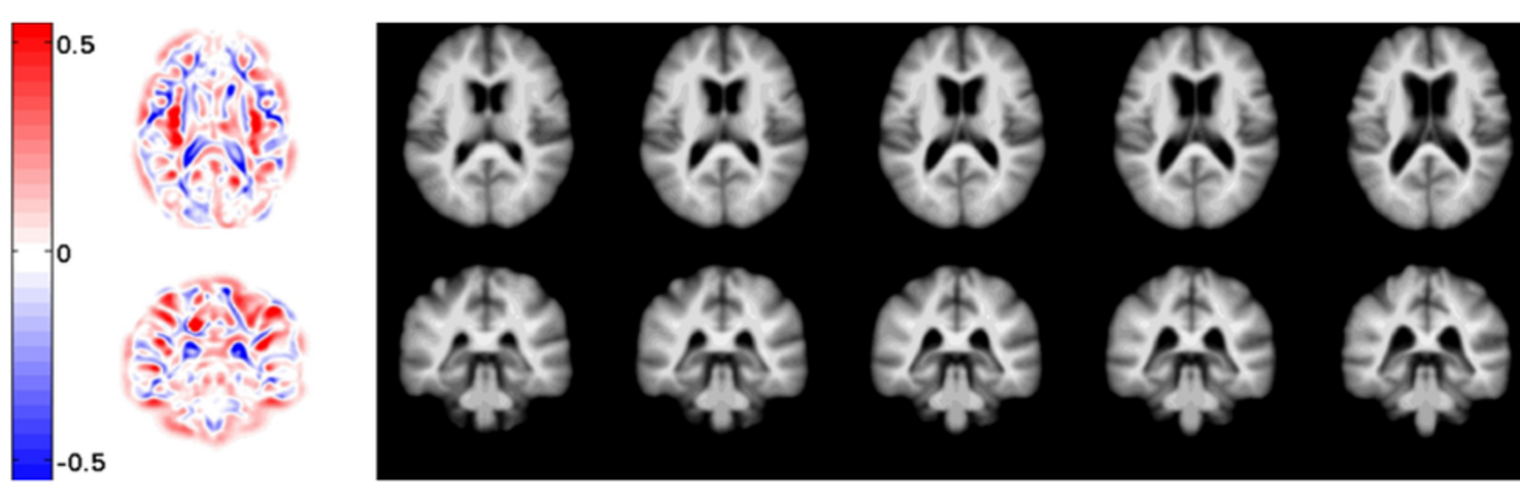
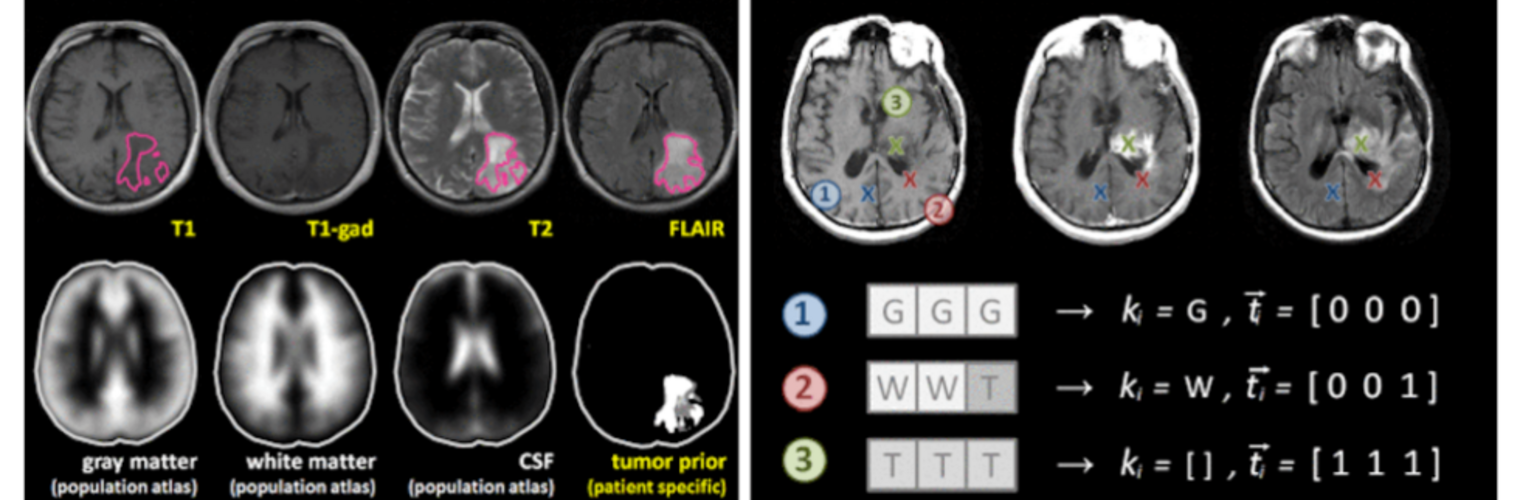
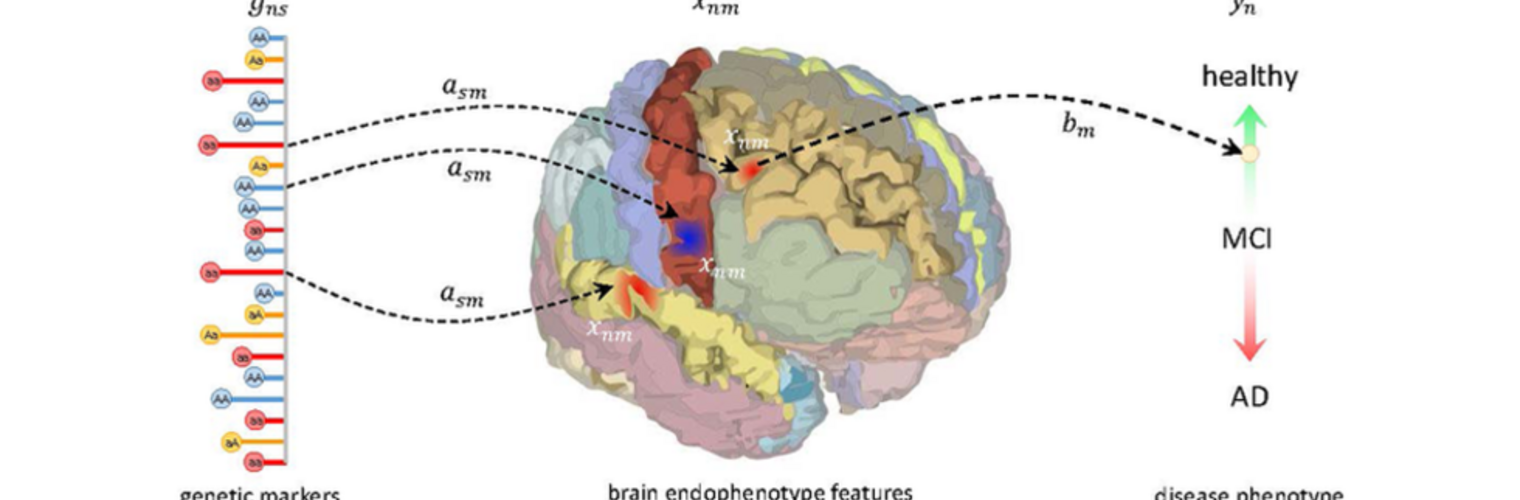
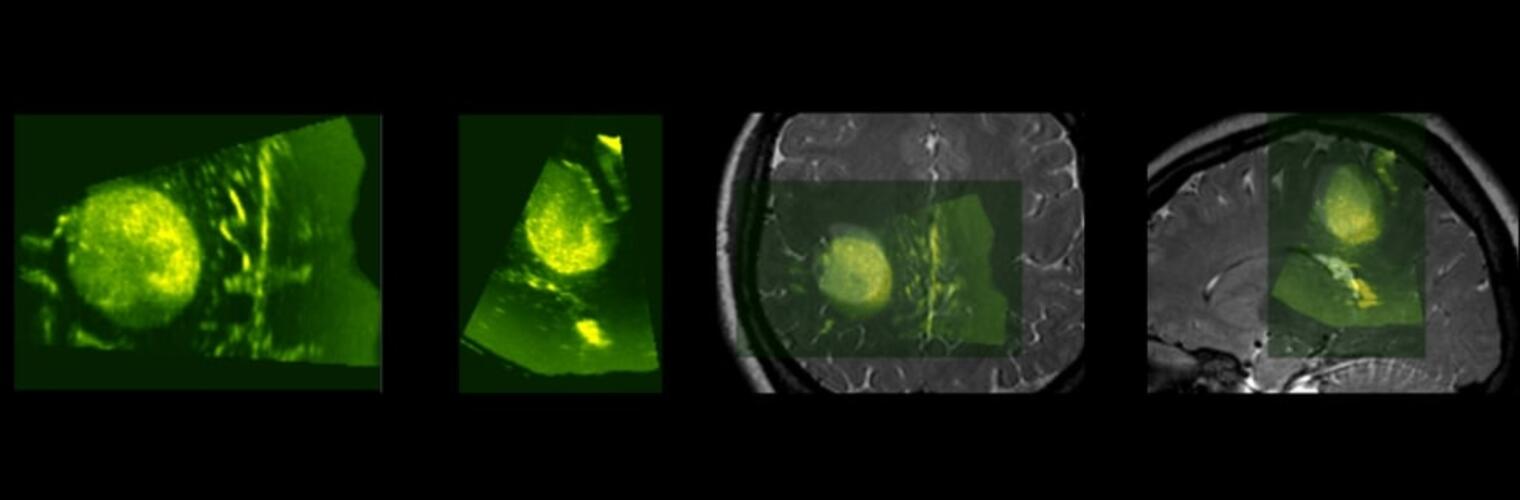
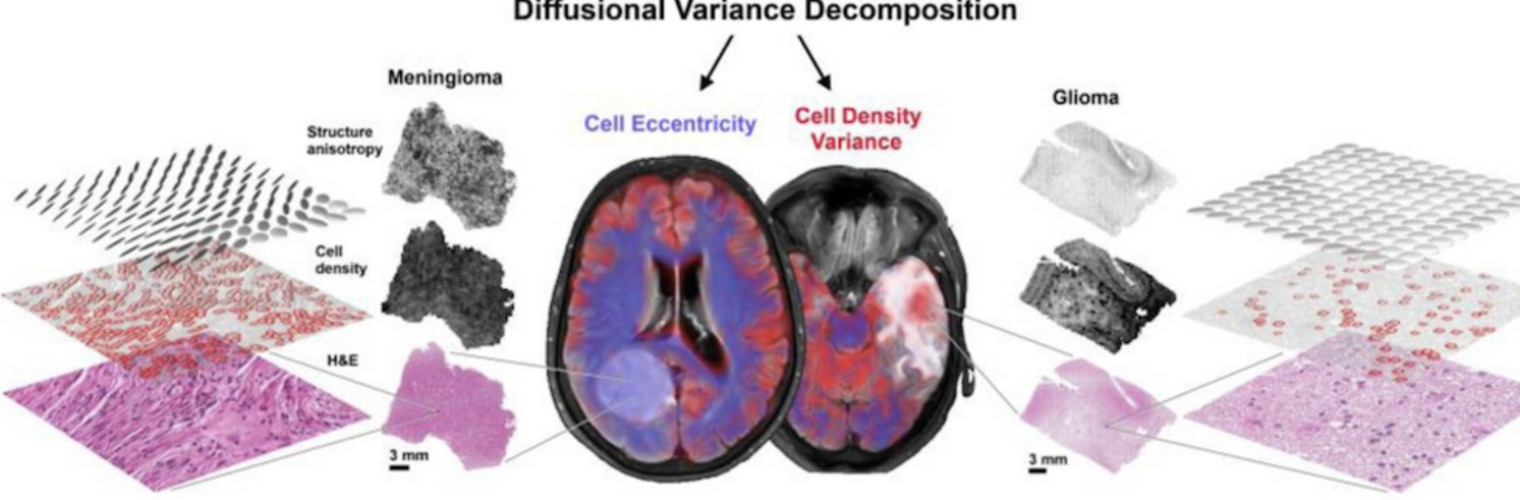
Filter exchange imaging (FEXI) is a double diffusion-encoding (DDE) sequence that is specifically sensitive to exchange between sites with different apparent diffusivities. FEXI uses a diffusion-encoding filtering block followed by a detection block at varying mixing times to map the exchange rate. Long mixing times enhance the sensitivity to exchange, but they pose challenges for imaging applications that require a stimulated echo sequence with crusher gradients. Thin imaging slices require strong crushers, which can introduce significant diffusion weighting and bias exchange rate estimates. Here, we treat the crushers as an additional encoding block and consider FEXI as a triple diffusion-encoding sequence. This allows the bias to be corrected in the case of multi-Gaussian diffusion, but not easily in the presence of restricted diffusion. Our approach addresses challenges in the presence of restricted diffusion and relies on the ability to independently gauge sensitivities to exchange and restricted diffusion for arbitrary gradient waveforms. It follows two principles: (i) the effects of crushers are included in the forward model using signal cumulant expansion; and (ii) timing parameters of diffusion gradients in filter and detection blocks are adjusted to maintain the same level of restriction encoding regardless of the mixing time. This results in the tuned exchange imaging (TEXI) protocol. The accuracy of exchange mapping with TEXI was assessed through Monte Carlo simulations in spheres of identical sizes and gamma-distributed sizes, and in parallel hexagonally packed cylinders. The simulations demonstrate that TEXI provides consistent exchange rates regardless of slice thickness and restriction size, even with strong crushers. However, the accuracy depends on b-values, mixing times, and restriction geometry. The constraints and limitations of TEXI are discussed, including suggestions for protocol adaptations. Further studies are needed to optimize the precision of TEXI and assess the approach experimentally in realistic, heterogeneous substrates.
Neuroimage Analysis Center
The Neuroimaging Analysis Center is a research and technology center with the mission of advancing the role of neuroimaging in health care. The ability to access huge cohorts of patient medical records and radiology data, the emergence of ever-more detailed imaging modalities, and the availability of unprecedented computer processing power marks the possibility for a new era in neuroimaging, disease understanding, and patient treatment. We are excited to present a national resource center with the goal of finding new ways of extracting disease characteristics from advanced imaging and computation, and to make these methods available to the larger medical community through a proven methodology of world-class research, open-source software, and extensive collaboration.
Our Sponsor

The NAC is a Biomedical Technology Resource Center supported by the National Institute of Biomedical Imaging and Bioengineering (NIBIB) (P41 EB015902). It was supported by the National Center for Research Resources (NCRR) (P41 RR13218) through December 2011.
Contact the Center Directors
|
|
Carl-Fredrik Westin, PhD Laboratory of Mathematics in Imaging Brigham and Women's Hospital 1249 Boylston St., Room 240 Boston, MA 02215 Phone: +1 617 525-6209 E-mail: westin at bwh.harvard.edu |
|
|
|
Ron Kikinis, MD Surgical Planning Laboratory Brigham and Women's Hospital 75 Francis St, L1 Room 050 Boston, MA 02115 Phone: +1 617 732-7389 E-mail: kikinis at bwh.harvard.edu |