
|
| Mike Halle, PhD Core PI |
Our Publications
The overarching goal of the Center Infrastructure Core, is to develop and maintain a standard software foundation to support the atlases developed by the other technology cores. The Atlas Infrastructure Core works closely with other NAC researchers to encourage the translation of research technologies and ideas into robust, shareable anatomical atlases, as well as with NAC's dissemination, outreach, and training activities. The work of this core is organized under the following specific aims.
- Develop tools to assist data authors and editors to name regions (both textual names and standardized names defined by controlled medical vocabularies), group regions into sets and hierarchies, and assign data and visual properties to these regions and groups.
- Develop tools for atlas editors to partition data authoring and editing tasks between two or more authors, preview the difference between the results, reconcile these differences, and merge the results.
- Develop an atlas-tailored revision control system for managing changes made both by individual content authors (Aim 1) and by atlas editors (Aim 2). This system will track changes in atlas image data and metadata and will integrate with tools for atlas release and distribution. We will release all tools developed as part of this infrastructure project as open source software licensed under the non-restrictive 3D Slicer License. We will use the NAC MRI Brain Atlas as an initial atlas dataset, while working with David Kennedy at University of Massachusetts Medical School to accommodate other datasets within our infrastructure.
Digital anatomy atlases are curated datasets consisting of imaging data, a set of labeled regions (often hand-edited) derived from the images, metadata and other parameters about those regions such as names or grouping information, and parameters that control how the atlas and its regions are rendered visually. Anatomy atlases are often the most distilled end product of an entire research effort consisting of many processes and algorithms. As such, atlases are extremely useful to other researchers both within a center such as ours and outside. Based on our experience in developing anatomy atlases over twenty years, these datasets have an extremely broad utility and appeal: we have received download requests from a broad spectrum of users, from physicians to device manufacturers to medical illustrators to members of the general public interested in human anatomy. In our Center, it has proved time-consuming and difficult to develop, organize, and maintain digital anatomy atlases. For example, for large atlases with hundreds of regions, authoring tools are either inadequate or simply do not exist to consistently name structures, group and organize them, and assign visual properties to them for display. Changes to properties and edits to the underlying regions of existing atlases are hard to manage, making it difficult to improve an atlas without introducing errors or artifacts. The problem of revision management is compounded when several people edit the atlas at once, and even more so when the atlas team includes experts physically located at other institutions. Using current tools, we have found it essentially impossible to have more than two people working on an atlas simultaneously. We are not aware of any existing software packages, either research or commercial, that are designed to meet the range of challenges posed by atlas creation. New infrastructure and tools are thus required to create richer, more accurate, and more useful atlases. The work of this core is organized under the following specific aims.
Featured Technologies
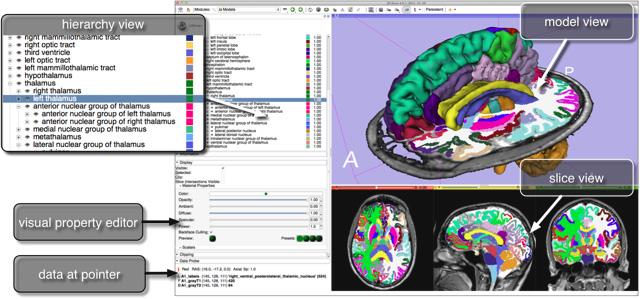
The NAC/CCA Brain Atlas consists of high quality multi-modal MR acquisitions and more than 150 segmented brain regions. These regions are cross-referenced with the RadLex and FMA ontologies and organized into a structural hierarchy. Pre-selected views of various anatomical systems are included in the Slicer version of the atlas.
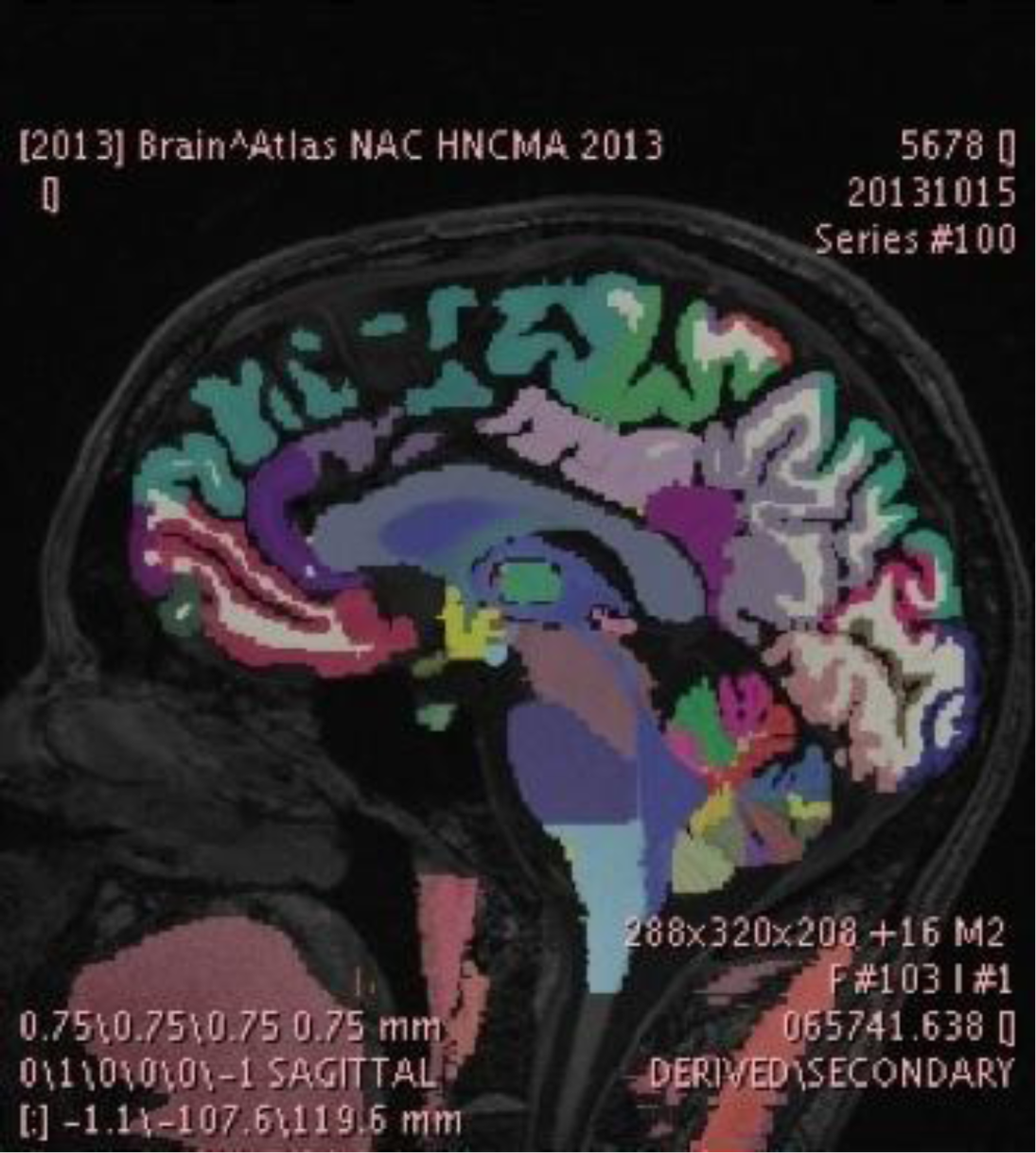
The SPL-PNL Brain Atlas label map displayed in the PixelMed tool developed by Dr. David Clunie. The atlas data was created and refined using a number of research tools, including FreeSurfer and 3D Slicer, and is made available on the 3D Slicer Data Store in Medical Reality Bundle (.mrb) format, which is an application-specific format of 3D Slicer. The data was converted by Dr. Clunie into the recently developed and standardized DICOM Segmentation Object Format to facilitate interoperability across research and community workstations as the Segmentation Object standard becomes widely adopted. We plan to work further on this topic in the coming year and integrate these standards based approaches. These algorithms will be used for a variety of clinical applications. The research will build on previous accomplishments in Atlas development that have included volumetric, geometric, structural, and functional atlases of anatomy.
Research Highlights
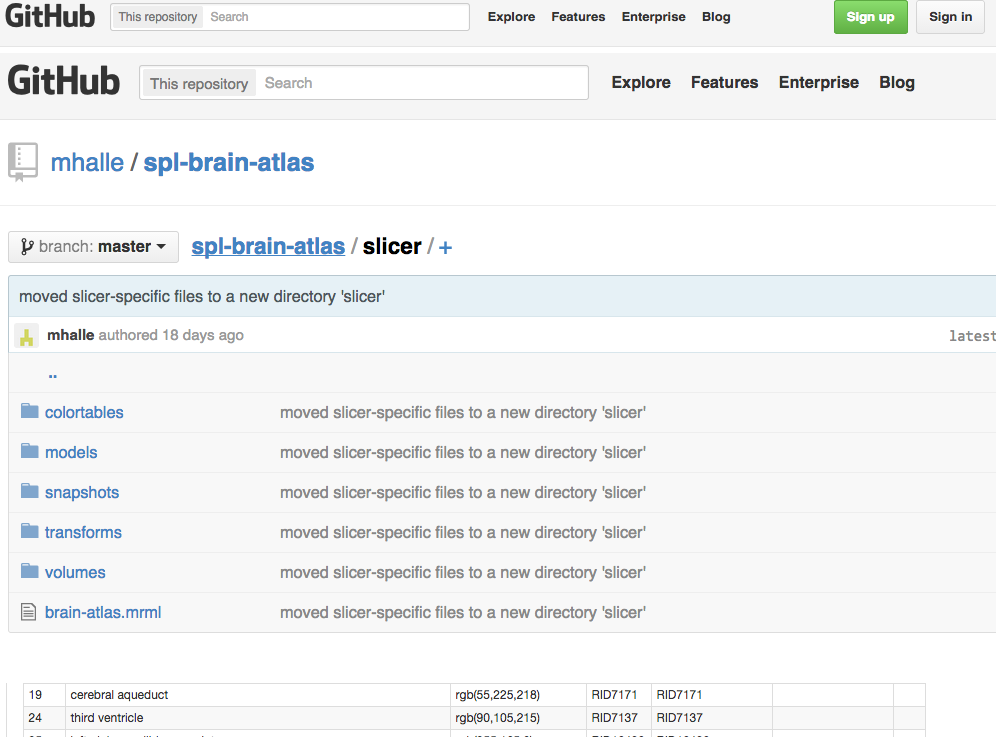
Figure 1: GitHub SPL/NAC-Brain Atlas. The NAC Brain Atlas was moved to GitHub to improve access for users and developers.
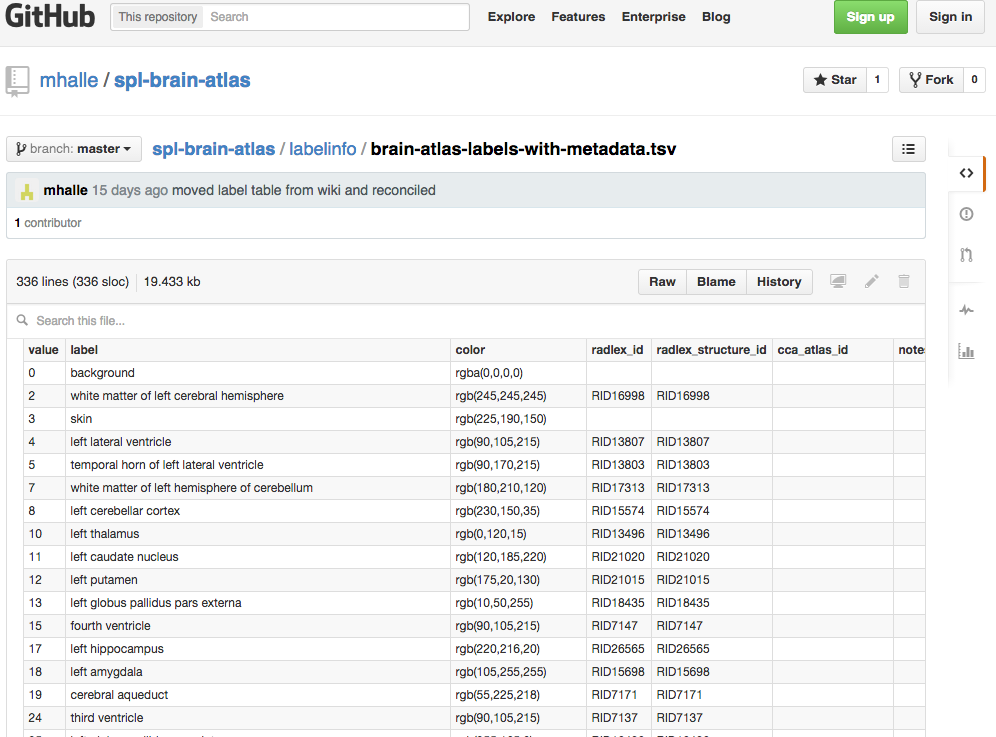
Figure 2: Labels-metadata. Taking advantage of tools provided by GitHub to make data more useful and accessible to other users, we converted the color lookup tables into more common tab-separated value files.
During the reporting interval, we have focused our attention on making the details of the process of atlas generation available to the larger community using tools we have refined for software development. In particular, we have created a new version of the NAC Brain Atlas and placed it on the collaborative software development site GitHub, where it can be found, copied, and downloaded by anyone.
Over the past few years, the use of GitHub and other similar sites has changed how collaborative software is developed by building community, openness, and accessibility from source code to version changes to compilation and distribution. GitHub and git, the underlying technology it uses for software revision control, are extremely well suited to serving multiple distributed teams working on text-like data. Management of “text-like data” is a critical aspect of git and GitHub’s design that presents potential challenges, in using it as we would like, to manage anatomy atlas data and metadata. Git maintains copies (or differences between copies) of all versions of every file under its control. Changes to software source code and other kinds of text are generally small and easily managed by git. However, changes to “non-text like” binary data, such as source radiographic image data or label maps, are usually handled naively as wholesale changes to files, losing the semantic meaning of the changes and potentially increasing the size of the data repository to unmanageable size.
Given the strong appeal of the widely used and understood development and collaborative infrastructure of GitHub, we have focused our efforts on bringing a sample atlas into a GitHub repository to understand its practical limitations, to make the existing atlas data fully available to other researchers, to begin building distributed tools to inspect and analyze the data within the repository, to establish a user and developer workflow using version-controlled data, and to otherwise adapt our approach to achieving this project’s specific aims.
Progress under Aims 1 and 2
Improving atlas data for GitHub
We have restructured the NAC Brain Atlas while moving it to the GitHub repository in order to improve its usefulness for users and developers (FIGURE 1). Previously, atlas data files were stored in a directory on a file system and typically accessed by a small group of people. Whenever the atlas was updated, changes were made to the three-dimensional models and hierarchical data and processed using 3D Slicer to create Slicer Scene Views. The collection of Scene Views was then packaged into a single slicer scene bundle called an MRB file.
This process produced a version of the atlas that was very convenient for Slicer users, and we will continue to offer an MRB file for use with Slicer going forward. However, this workflow meant that other users who might want direct access to segmentation files or 3D models of brain anatomy had to understand details about 3D Slicer and the implementation of its internal file structures. In the new GitHub-based workflow, atlas data is stored more categorically, with source medical images, segmentation label files, and 3D models. Object metadata, such as names and display colors, are organized in separate folders to make them easier to find. Slicer-specific files, such as MRML scene descriptions and color lookup tables, are stored separately from the generic atlas data as often as possible. We are currently working on a separate repository of tools that will contain software that will convert generic information into Slicer files and bundle all the atlas data into a single MRB file.
Progress under Aims 3
Using GitHub’s User Interface Features
We are also taking advantage of tools provided by GitHub to make data more useful and accessible to other users. For example, we have converted our color lookup tables, which contain object names and colors, into more common tab-separated value files. These files can be previewed and searched in GitHub’s web interface (FIGURE 2). These files can also be downloaded straight into programs such as Microsoft Excel, as well as contain additional structural metadata such as linkage to other ontologies and formal naming schemes. A small software tool then converts these files into Slicer lookup tables.
During the rest of the reporting period, we will produce versions of our anatomy models in STL format, which is widely used in the 3D printing community and natively viewable in GitHub’s online data viewing tools. Development of these tools that are useful to the end user help satisfy our goals in Specific Aim 1. Understanding the implications of version controlled atlas data Now that the NAC Brain Atlas GitHub repository exists, we can explore the issues related to the mechanics of managing binary data as required by Specific Aim 3. We have already made some observations. First, the source MRI imaging upon which the atlas is based is essentially never edited and very infrequently augmented. This fact is critical because we have only the most limited ability to capture meaningful information about the differences between one image version and another. As a result, we must store each version of these images as if they were completely new, which can be very expensive from a storage point of view. This fact is especially true in distributed revision control systems such as git, which sends all versions of a file to every developer or users by default. The fact that the largest data in the atlas is almost immutable means that the atlas is not likely prone to this problem, though we will monitor it. Atlas metadata, such as colors, names, and hierarchal descriptions, which are relatively small and text-based, are fairly easy to represent and understand as they change from version to version. We will focus on ways to edit this data, with the underlying GitHub repository being the only data source required (so no files will need to be downloaded and processed locally).
Finally, the label map images that describe the different structural regions in the atlas provide the biggest challenge for version control. These maps are smaller and more compressible than the source images, but are binary and larger in size compared to the text-based atlas metadata. They are also likely to represent the most edited element of the atlas, so capturing meaningful differences between versions is both more important and more difficult than with the other types of atlas data. We will continue to study this problem as project work continues.
