
|
| Steve Pieper, PhD Core PI |
Our Publications
The role of the Slicer Core, is to facilitate the practical translation of NAC technologies to a user accessible platform in support of internal NAC research, clinical application, and broader dissemination. These goals are accomplished through software innovation and the creation and maintenance of software for 3D Slicer (a.k.a. Slicer), a package which has grown over the last 10 years from a graduate student project within NAC to a leading tool in the image analysis field. Slicer has a strong network of contributing authors supported by a number of funding mechanisms including the National Alliance for Medical Image Computing (NA-MIC) and the National Center for Image Guided Therapy (NCIGT). NCIGT also provides the driving biomedical project (DBP) for this core. Each of our partners brings a unique set of driving projects and investigators that strengthen the underlying Slicer platform. The NCI Quantitative Imaging Network (QIN) and the Ontario SparKit efforts, both of which are service collaborations of NAC, provide additional motivating use cases and development synergy. Within the NAC research community, creation of Slicer-compatible code is a recognized sign that ideas have matured from initial concept to a form suitable for users to test with their own data. Clinical domain experts are the key group of users for Slicer. They provide both the problem definitions and the working context in which new ideas and approaches can be evaluated and improved. These efforts result in code modules in Slicer that form the basis for outreach efforts. In this way, the research community can benefit from both the original published method and the embodiment of the concept within a working tool. The work of this core is organized around the following specific aims.
- Refine Slicer as a research platform for creation and dissemination of NAC technologies.
- Adapt Slicer to rapidly integrate, analyze, and visualize multimodal data.
- Define and apply Slicer CaseHub tools to neurosurgery in AMIGO.
In addition to providing leadership and participation in the 3D Slicer community and other national and international efforts (aim 1), the core is creating a new set of user-steered tools to permit rapid analysis and visualization of multimodal data driven by the needs of its DBP (aim 2), as well as a new “CaseHub” framework to manage sequentially acquired scans and other data from ongoing clinical scenarios. The idea is to enable clinical experts to review and understand the evolving patient state and determine next treatment steps in a dynamic and time constrained environment, where critical decisions must be made within a 10-15 minute window of time (aim 3). The target of this work is the NCIGT's Advanced Multimodality Image Guided Operating (AMIGO) Suite. AMIGO is the clinical translational testbed of NCIGT. In AMIGO, real-time anatomical imaging modalities, such as X-ray and ultrasound, can be combined with cross-sectional digital imaging systems including CT, MRI, and PET. Molecular image-guided therapies are investigated with multiple molecular probes, such as PET, optical imaging, and targeted mass spectrometry, to increase the sensitivity and specificity of cancer detection. The application of these tools and data management framework is expected to improve the definition of tumor margins to enable more complete excision and/or thermal ablation. As illustrated in the three Featured Technologies below, the improved Slicer user-steered tools and CaseHub server technology have continued to evolve and have taken the form of (1) advanced Slicer-based image analysis tools, (2) custom software for MR and ultrasound data integration in neurosurgery, and (3) evaluations of web applications as a platform for NAC research.
Featured Technologies
3D Slicer Platform
Figure 1 shows just some of the many projects building on the 3D Slicer platform and the bullets below include links and short descriptions of some of the notable developments of the past year involving and building on NAC technology.
- Segmentations / Segment Editor - powerful new infrastructure to represent and operate on image segmentations including natively 3D interactions.
- Quantitative Imaging / DICOM - greatly improved standards-based data representation, validated in the context of connectathons at RSNA
- SlicerRadiomics - researcher-friendly tools for image feature calculation.
- SlicerDMRI - world-class diffusion analysis and tractography tools.
- SlicerCIP - chest imaging platform to quantify airways and related structures.
- SlicerHeart - new 4D ultrasound processing and cardiac analysis tools.
- SlicerSALT - a comprehensive medical image shape analysis package.
- SlicerCMF - custom tools and distribution for dental and craniomaxillofacial surgery.
- SlicerRT - extensive infrastructure for radiotherapy research.
- SlicerIGT - custom modules to build fully functional image guided surgery prototypes.
- SlicerPathology - mixing 3D Slicer with cloud databases for terascale microscopy analysis.
Full documentation of the 3D Slicer ecosystem is always available at http://slicer.org.

Figure 1. A sampling of 3D Slicer activities.
Slicer-based Image Analysis
Hierarchical Segmentation of Brain Pathology Images
Extracting nuclei is one of the most actively studied topics in digital pathology research. It is essential in surgical planning and may be combined with mass spectrometry data during surgical procedures. Most of the studies directly search the nuclei (or seeds for the nuclei) from the finest resolution available. While the richest information has been utilized by such approaches, it is sometimes difficult to address the heterogeneity of nuclei in different tissues. In our recent work, we propose a hierarchical approach, which starts from the lower resolution level and adaptively adjusts the parameters while progressing into finer and finer resolution. This approach employs the Fast GrowCut algorithm as a key part of the pipeline. Fast GrowCut was developed as part of our NAC-sponsored research. This work is embodied in our SlicerPathology extension as illustrated in Figure 2 and can be seen in a video demonstration online.
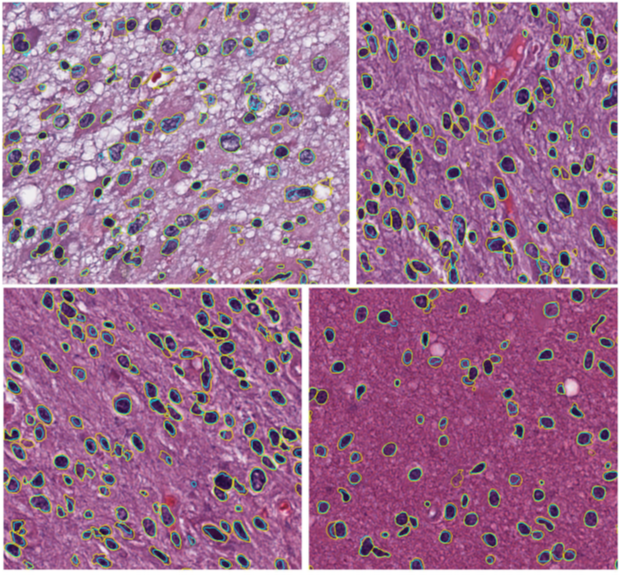
Figure 2. Four example brain Images: Yellow manual; cyan algorithmic. Taken from 15 TCGA brain images about 700 by 700 with resolution 0.25 µm/pixel.
MR-Ultrasound Fusion for Neurosurgery
NAC has developed the AmigoDataCollection module as a concrete implementation of the CaseHub architecture to integrate multiple modes of intraprocedural image data into a unified data space for visualization and analysis. As shown in the figure, the custom module provides push-button data acquisition options for volumetric ultrasound acquisition, stereo image capture, and recording of the position and orientation of tracked surgical instruments.
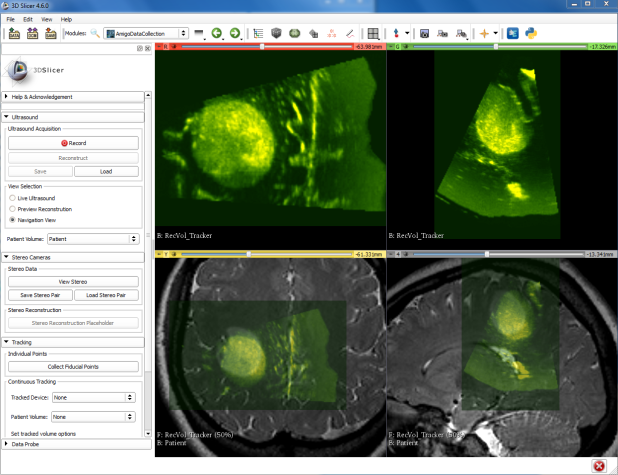
Within 3D Slicer this data can be interactively reviewed, as demonstrated in this video showing two timepoints of intraprocedural ultrasounds superimposed on pre-procedure MR images.
Web Applications for Neuroimage Analysis
NAC continues to innovate in the area of software infrastructure technologies for neuroimage analysis, and in the context of intraprocedural imaging we seek easily deployable yet high performance computing environments. To this end, we have looked at the question of GPU accelerated image computing that can run in the latest breed of conventional web browsers. Specifically we investigated the use of WebGL2 technology, which is now standard in Google Chrome and Mozilla Firefox browsers, as a platform for image processing operations. WebGL2 is a Khronos industry standard set of JavaScript bindings to the OpenGLES 3.0 functionality that is ubiquitous in electronic devices from smart TVs through mobile phones and desktop computers and advanced workstations. The same WebGL2 code that runs on the Chrome browser on an Android smartphone can also leverage the computing power of the latest generation of workstation graphics cards, thus supporting a wide range of use scenarios with a single code base. To evaluate the functionality we implemented and tested two important pieces of functionality as demonstrated in the video linked here: (1) we load and correctly display MR neuroimaging data in native DICOM format, and (2) we apply compute intensive 3D nonlinear image filtering at interactive rates. The image filter in question, the bilateral filter, is implemented as a multithreaded filter in ITK and consumes the full compute resources of an 8 core Xeon E5-2620 @2.1 GHz for over 8 minutes. In contrast, the same calculation is performed in approximately 2 seconds on an Nvidia GTX 1080 graphics card when implemented in GLSL shader code on WebGL2, a performance improvement of over 200x. Based on these results we are continuing to develop our WebGL2 based computing framework.
